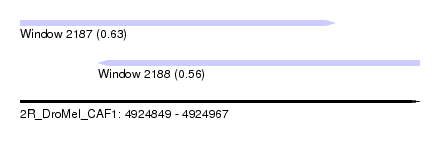
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,924,849 – 4,924,967 |
| Length | 118 |
| Max. P | 0.632732 |

| Location | 4,924,849 – 4,924,942 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 87.61 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.77 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4924849 93 + 20766785 AUACGUACGUACUUUCCUUCGCUGGCUUUUCGCAAUUUGUUUAUGUGACCAUCAAUUUGUGAGGCUCCACAGACAC-UUUGUGUUUGCCGGCCA .(((....))).........((((((....(((((..(((((.((.((((.(((.....))))).)))).))))).-.)))))...)))))).. ( -26.80) >DroSec_CAF1 13697 87 + 1 ------ACGUACUUUCGUUCGCUGGCUUUUCGCAAUUUGUUUAUGUGACCAUCAAUUUGUGAGGCUCCACAGACAC-UUUGUGUUUGCCGGCCA ------(((......)))..((((((....(((((..(((((.((.((((.(((.....))))).)))).))))).-.)))))...)))))).. ( -26.90) >DroSim_CAF1 13865 87 + 1 ------ACGUACUUUUUUUCGCUGGUCUUUCGCAAUUUGUUUAUGUGACCAUCAAUUUGUGAGGCUCCACAGACAC-UUUGUGUUUGCCGGCCA ------..............((((((....(((((..(((((.((.((((.(((.....))))).)))).))))).-.)))))...)))))).. ( -24.30) >DroEre_CAF1 11952 87 + 1 ------ACGUACUUUUGUUCGCUGGUUUUUCGCAGUUUGUUUAUGUGACCAUCAAUUUGUGAGGCUCCACAGACAC-UUUGUGUUUGCCGGCCA ------..............((((((....(((((..(((((.((.((((.(((.....))))).)))).))))).-.)))))...)))))).. ( -24.70) >DroYak_CAF1 13947 86 + 1 -------CGUACAUUCGUUCGCUGGGUUUUCGCAAUUUGUUUAUGUGACCAUCAAUUUGUGAGGCUCCACAGACAC-UUUGUGUUUGCCGGCCA -------.............(((((.....(((((..(((((.((.((((.(((.....))))).)))).))))).-.)))))....))))).. ( -22.30) >DroAna_CAF1 10797 83 + 1 ------ACACACCAUCAGCCAC-----AUUCGCAAUUUGUUUAUGCGACCAUCAAUUCGUGAAGCUCCACAGACACACUUGUGUUUGCCGGCCA ------...........(((..-----..(((((.........))))).....................(((((((....)))))))..))).. ( -17.10) >consensus ______ACGUACUUUCGUUCGCUGGCUUUUCGCAAUUUGUUUAUGUGACCAUCAAUUUGUGAGGCUCCACAGACAC_UUUGUGUUUGCCGGCCA ....................((((((....(((((..(((((.((.((((.(((.....))))).)))).)))))...)))))...)))))).. (-18.85 = -19.77 + 0.92)

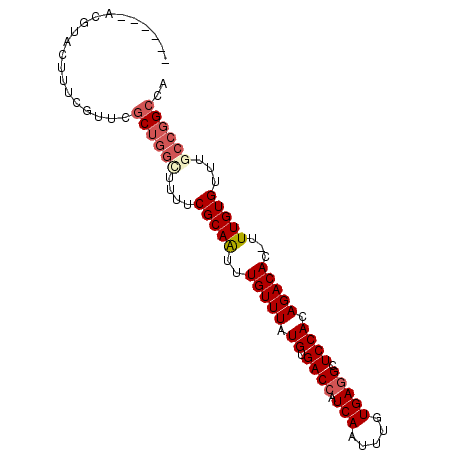
| Location | 4,924,872 – 4,924,967 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -15.41 |
| Energy contribution | -16.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4924872 95 - 20766785 AU------CUUGGCCAUUUUAUCGAUUUGCU-----UGGCCGGCAAACACAAA-GUGUCUGUGGAGCCUCACAAAUUGAUGGUCACAUAAACAAAUUGCGAAAAGCC ..------..((((((.....((((((((..-----.((((.(((.((((...-)))).))).).)))...))))))))))))))............((.....)). ( -27.20) >DroPse_CAF1 11817 103 - 1 AUUUGUUACCUCUCGAUUUUAUUGAUUUGCUUGGCUUGGCCGGCAAACACAAA-AGUGCUCCACCGCCUCCAAAUUUGGCGGUCACGUAAACAAAU---GAAAAUCG .............(((((((.(((.((((((.(((...)))))))))......-.(((....((((((.........))))))))).....)))..---.))))))) ( -26.30) >DroSim_CAF1 13882 95 - 1 AU------CUUGGCCAUUUUAUCGAUUUGCU-----UGGCCGGCAAACACAAA-GUGUCUGUGGAGCCUCACAAAUUGAUGGUCACAUAAACAAAUUGCGAAAGACC ..------..((((((.....((((((((..-----.((((.(((.((((...-)))).))).).)))...)))))))))))))).............(....)... ( -27.30) >DroEre_CAF1 11969 95 - 1 AU------CUUGGCCAUUUUAUCGAUUUGCU-----UGGCCGGCAAACACAAA-GUGUCUGUGGAGCCUCACAAAUUGAUGGUCACAUAAACAAACUGCGAAAAACC ..------..((((((.....((((((((..-----.((((.(((.((((...-)))).))).).)))...))))))))))))))...................... ( -26.40) >DroYak_CAF1 13963 95 - 1 AU------CUUGGCCAUUUUAUCGAUUUGCU-----UGGCCGGCAAACACAAA-GUGUCUGUGGAGCCUCACAAAUUGAUGGUCACAUAAACAAAUUGCGAAAACCC ..------..((((((.....((((((((..-----.((((.(((.((((...-)))).))).).)))...))))))))))))))...................... ( -26.40) >DroAna_CAF1 10813 92 - 1 AG------CUCCGCUAUUUUAUUGAUUUGCU-----UGGCCGGCAAACACAAGUGUGUCUGUGGAGCUUCACGAAUUGAUGGUCGCAUAAACAAAUUGCGAAU---- ..------..........((((..(((((..-----.((((.(((.((((....)))).))).).)))...)))))..))))(((((.........)))))..---- ( -25.60) >consensus AU______CUUGGCCAUUUUAUCGAUUUGCU_____UGGCCGGCAAACACAAA_GUGUCUGUGGAGCCUCACAAAUUGAUGGUCACAUAAACAAAUUGCGAAAAACC ..........((((((.....((((((((........((((.(((.((((....)))).))).).)))...))))))))))))))...................... (-15.41 = -16.42 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:22 2006