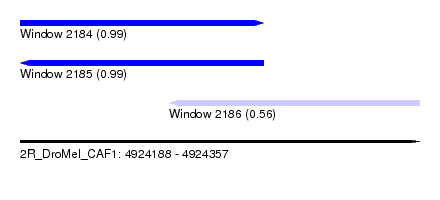
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,924,188 – 4,924,357 |
| Length | 169 |
| Max. P | 0.993777 |

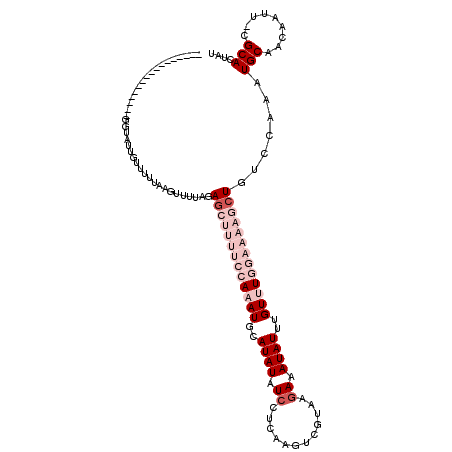
| Location | 4,924,188 – 4,924,291 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.03 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -7.42 |
| Energy contribution | -11.02 |
| Covariance contribution | 3.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.30 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4924188 103 + 20766785 -----------------GGAAUUGUUUUCACGUUUUCGAGCUUUUCCAAAUUCAUAUAUCCUAAAGUCGUAAGAAAUAUUUGUUUGGAAAAGCUGUCCAAAUGCAAGCAAUUCGCACUAU -----------------.(((((((((.((((....))(((((((((((((..((((.((.((......)).)).))))..))))))))))))).......)).)))))))))....... ( -29.00) >DroSec_CAF1 13034 103 + 1 -----------------AGUAUUGUUUAUAAUUUUUAGAACUUUUCCAAAUGCAUAUAUCCUCAAGUCGUAGGAAAUAUUUGUUUGGAAAAGCUGUCCAAAUGCAACAAUUCCGCACUAU -----------------.((((((((....((((.(((..((((((((((((.((((.(((((.....).)))).)))).)))))))))))))))...))))..))))))...))..... ( -21.80) >DroSim_CAF1 13200 103 + 1 -----------------AGUAUUGUUUAUAAGUUUUAGAGCUUUUCCAAAUGCAUAUAUCCUCAAGUCGUAGGAAAUAUUUGUUUGGAAAAGCUAUCCAAAUGCAACAAUUCCGCACUAU -----------------.((((((((.....((((...((((((((((((((.((((.(((((.....).)))).)))).))))))))))))))....))))..))))))...))..... ( -27.00) >DroEre_CAF1 11262 113 + 1 UGCUAUAAUCUGAAUUUGGUGCUGCUCUUAAGGUAUAGAGCUUUUUCAUAUGCAUAUAUGCCAAAGUCGUAAAAAAUAUGUGUUUU------CUGCCCAAGUGCAGCAACU-CGCACUAA (((......(((.((((((.((.(((((........)))))..........((((((((................))))))))...------..)))))))).))).....-.))).... ( -24.79) >DroYak_CAF1 13258 113 + 1 GGCUAGAAUCUAAAUUUGGCAUUGUUUUCAAGUUAUAGAGCUUUUUCACAUGCAUAUAUCCUCAAGUCGUAAGAAAUAUUUGUUUU------CUGCCCAAGUGCAGCAAUU-CGCACUAU (((.((((..(((((...((((.((....(((((....)))))....)))))).............((....))...)))))..))------)))))..(((((.......-.))))).. ( -22.80) >consensus _________________GGUAUUGUUUUUAAGUUUUAGAGCUUUUCCAAAUGCAUAUAUCCUCAAGUCGUAAGAAAUAUUUGUUUGGAAAAGCUGUCCAAAUGCAACAAUU_CGCACUAU ......................................(((((((((((((..((((.((............)).))))..))))))))))))).......(((.........))).... ( -7.42 = -11.02 + 3.60)

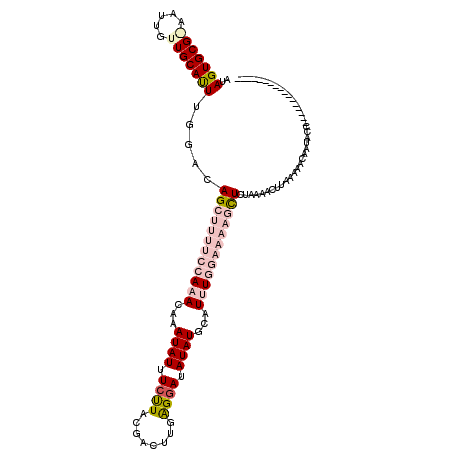
| Location | 4,924,188 – 4,924,291 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.03 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -13.66 |
| Energy contribution | -16.58 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4924188 103 - 20766785 AUAGUGCGAAUUGCUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCUUACGACUUUAGGAUAUAUGAAUUUGGAAAAGCUCGAAAACGUGAAAACAAUUCC----------------- ..(((((((.....))))))).(((.((((((((((((...((((.(((((......))))).))))...)))))))))))).......((....))...)))----------------- ( -28.40) >DroSec_CAF1 13034 103 - 1 AUAGUGCGGAAUUGUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCCUACGACUUGAGGAUAUAUGCAUUUGGAAAAGUUCUAAAAAUUAUAAACAAUACU----------------- .......((.((((((...((((((..(((((((((((...((((.((((........)))).))))...))))))))))))))))).......)))))).))----------------- ( -26.10) >DroSim_CAF1 13200 103 - 1 AUAGUGCGGAAUUGUUGCAUUUGGAUAGCUUUUCCAAACAAAUAUUUCCUACGACUUGAGGAUAUAUGCAUUUGGAAAAGCUCUAAAACUUAUAAACAAUACU----------------- .......((.((((((...((((((..(((((((((((...((((.((((........)))).))))...))))))))))))))))).......)))))).))----------------- ( -27.50) >DroEre_CAF1 11262 113 - 1 UUAGUGCG-AGUUGCUGCACUUGGGCAG------AAAACACAUAUUUUUUACGACUUUGGCAUAUAUGCAUAUGAAAAAGCUCUAUACCUUAAGAGCAGCACCAAAUUCAGAUUAUAGCA .......(-(((((((((.((((((.((------((((......))))))..((((((..(((((....)))))..)))).)).....)))))).))))))....))))........... ( -23.80) >DroYak_CAF1 13258 113 - 1 AUAGUGCG-AAUUGCUGCACUUGGGCAG------AAAACAAAUAUUUCUUACGACUUGAGGAUAUAUGCAUGUGAAAAAGCUCUAUAACUUGAAAACAAUGCCAAAUUUAGAUUCUAGCC ..((((((-......))))))..(((((------((.....((((((((((.....)))))).))))((((((....(((........)))....)).))))..........)))).))) ( -20.30) >consensus AUAGUGCG_AAUUGUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCUUACGACUUGAGGAUAUAUGCAUUUGGAAAAGCUCUAAAACUUAAAAACAAUACC_________________ ..(((((((.....))))))).....((((((((((((...((((.((((........)))).))))...))))))))))))...................................... (-13.66 = -16.58 + 2.92)

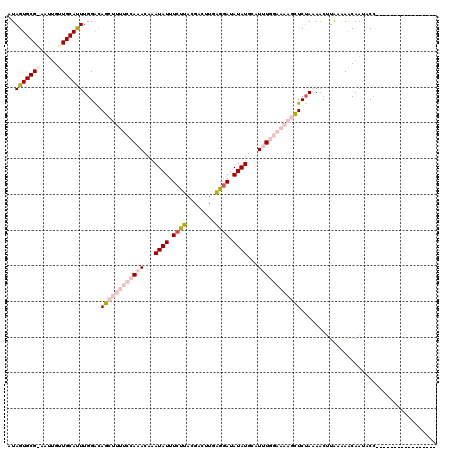
| Location | 4,924,251 – 4,924,357 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.16 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4924251 106 - 20766785 CACAAAAUUUCUGAGUGCUUACGGCGUAUAAUU-UUGCACACUUUUUUGGGCAAUUUGCAAAUGUUUAUAGUGCGAAUUGCUUGCAUUUGGACAGCUUUUCCAAACA .........((..(((((.....(((.......-.)))..........((((((((((((...........)))))))))))))))))..))............... ( -25.50) >DroSec_CAF1 13097 105 - 1 CACAG-AUUUCUGAGUGCUUUCGGCGUAUAAUU-UUGCCCACUUUUUUGGGCAAUUUGCAAAUGUUUAUAGUGCGGAAUUGUUGCAUUUGGACAGCUUUUCCAAACA ..(((-....))).(..((.(..((((...(..-(((((((......)))))))..)....))))..).))..)((((..((((........))))..))))..... ( -29.80) >DroSim_CAF1 13263 105 - 1 CACAG-AUUUCUGAGUGCUUUCGGCGUAUAAUU-UUGCACACUUUUUUGGGCAAUUUGCAAAUGUUUAUAGUGCGGAAUUGUUGCAUUUGGAUAGCUUUUCCAAACA ..(((-....)))..........(((.((((((-((((((.((.....))((.....))...........))))))))))))))).((((((.......)))))).. ( -25.30) >DroEre_CAF1 11342 99 - 1 CAUAC-AAUGCUGAGUGCUUUCGGCGUAAAAUUUUUGCUCAUUCUUUUGGGCAAUUUGUAAAUGUUUUUAGUGCG-AGUUGCUGCACUUGGGCAG------AAAACA ..(((-(((((((((....)))))))).......(((((((......)))))))..))))..(((((((.((.((-(((......))))).)).)------)))))) ( -31.10) >DroYak_CAF1 13338 98 - 1 CAGAG-AAUUCUGAGUGCUUUCGGCGUUUAAUU-UUGCACACUUUUUUGGGCAAUUUGUAAAUGUUUAUAGUGCG-AAUUGCUGCACUUGGGCAG------AAAACA .....-...((..(((((.....(((.......-.)))...........(((((((((((...........))))-))))))))))))..))...------...... ( -24.60) >consensus CACAG_AUUUCUGAGUGCUUUCGGCGUAUAAUU_UUGCACACUUUUUUGGGCAAUUUGCAAAUGUUUAUAGUGCG_AAUUGUUGCAUUUGGACAGCUUUUCCAAACA .........((..(((((....((((.........))).).........(((((((((((...........)))).))))))))))))..))............... (-16.90 = -16.74 + -0.16)

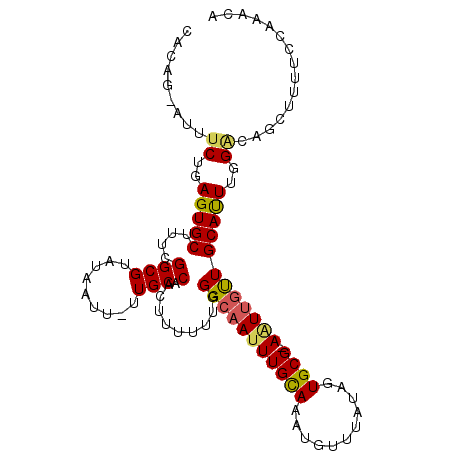
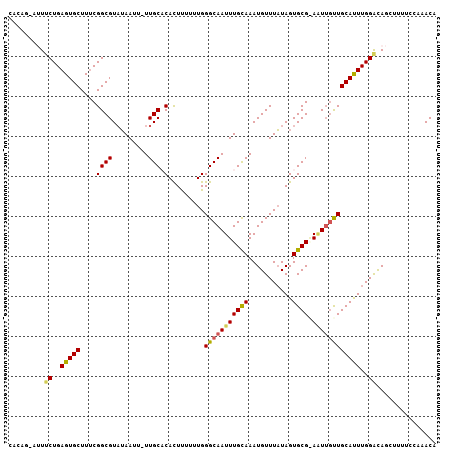
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:20 2006