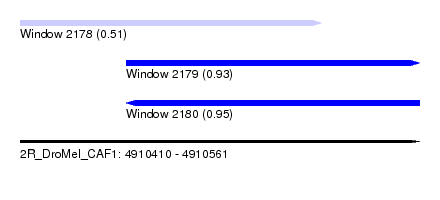
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,910,410 – 4,910,561 |
| Length | 151 |
| Max. P | 0.949451 |

| Location | 4,910,410 – 4,910,524 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.54 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4910410 114 + 20766785 CCACCAUGAUCUUUUUGAAGUUGUACAGACUCAUCUUGGCAGG-AUUUGUUUAUUUUCCACGCGU--C-G-AAAGGAAAAUAAAUGCGGAAUCGAAAGCAAAAACACCACGCGGUGUCA ...(((.(((...((((........))))...))).)))...(-(((((((((((((((......--.-.-...)))))))))))...)))))..........(((((....))))).. ( -22.50) >DroVir_CAF1 75493 116 + 1 CCACCAUUAUCUUUUUAAAGUUGUACAAACUCAUUUUUGUACUUAUUUAUUUAUUUUCCACGGCAA-A-UUAAAGGAAAAUAAAUG-GGAAUCGCUUGCAAAAACACCACGCGGUGUCU ............((((.((((.(((((((......)))))))...((((((((((((((.......-.-.....))))))))))))-))....))))..))))(((((....))))).. ( -24.12) >DroGri_CAF1 69581 115 + 1 CCACCAUUAUCUUUUUAAAGUUGUACAAACUCAUUUUCGCAGUUAUUUAUUUAUUUUCCAAGUUAA-U-C-UAAGGAAAAUAAAGG-GAAAUCGCUAACAAAUACACCACGCGGUGCCU .((((.............((((.....)))).........(((.((((.((((((((((.......-.-.-...))))))))))..-.)))).)))................))))... ( -18.40) >DroWil_CAF1 60186 117 + 1 CCACCAUUAUCUUUUUAAAAUUGUACAAACUCAUUUUUGUAGAAAUUUAUUUAUUUUCCAAGCCAAUU-A-AACGGAAAAUAAAUGAAAUGUGCAAAGCAAUAACACCACGCGGUGCUA .((((...............(((((((..((((....)).))...((((((((((((((.........-.-...)))))))))))))).))))))).((...........))))))... ( -25.42) >DroMoj_CAF1 93477 116 + 1 CCACCAUGAUCUUUUUAAAGUUGUACAAACUCAUUUUUGUGGCUAUUUAUUUAUUUUCCACGGCAA-A-A-AAAGGAAAAUAAAAGAGGAAUCGCUAGCAUAAACACCACGCGGUGUUC .......(((((((((..(((..((((((......)))))))))......(((((((((.......-.-.-...))))))))))))))).))).........((((((....)))))). ( -21.90) >DroPer_CAF1 37331 114 + 1 CCACCAUUAUCUUCUUAAAGUUGUACAAACUCAUUUUGUAUGG-UUUUAUUUAUUUUCCACGCGU--AAA-AAAGGAAAAUAAACGC-GAAUCUCAAGCAAAAACACCACGCGGUGCCU .((((..........(((((..(((((((.....)))))))..-)))))((((((((((......--...-...)))))))))).((-(....................)))))))... ( -23.25) >consensus CCACCAUUAUCUUUUUAAAGUUGUACAAACUCAUUUUUGUAGGUAUUUAUUUAUUUUCCACGCCAA_A_A_AAAGGAAAAUAAAUG_GGAAUCGCAAGCAAAAACACCACGCGGUGCCU .((((.............((((.....))))..................((((((((((...............))))))))))............................))))... (-13.52 = -13.54 + 0.03)

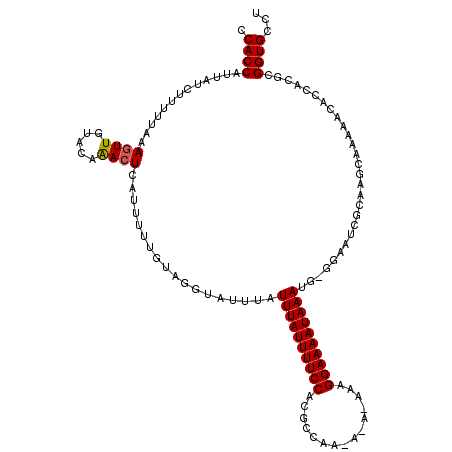
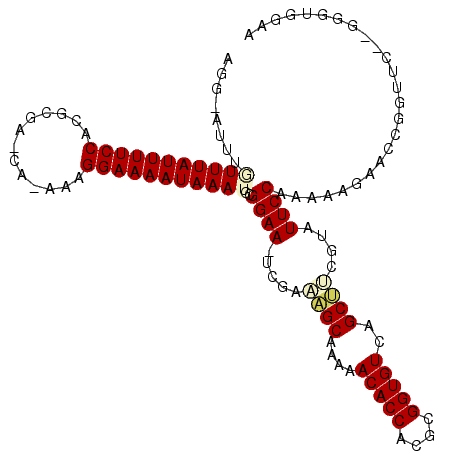
| Location | 4,910,450 – 4,910,561 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4910450 111 + 20766785 AGG-AUUUGUUUAUUUUCCACGCGU-CG-AAAGGAAAAUAAAUGCGGAA-UCGAAAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACCGGUUCUCGGGCGGAA ...-....(((((((((((......-..-...)))))))))))((((((-(((((.((....(((((....)))))..)))))).)))))........(((.....))))).... ( -32.30) >DroVir_CAF1 75533 106 + 1 ACUUAUUUAUUUAUUUUCCACGGCAAAUUAAAGGAAAAUAAAUG-GGAA-UCGCUUGCAAAAACACCACGCGGUGUCUGCAACAAAUUCCAAAAAGAGCUGAUAU------AGA- .(((...((((((((((((.............))))))))))))-((((-(...(((((...(((((....))))).)))))...)))))...))).........------...- ( -26.52) >DroSim_CAF1 62290 111 + 1 AGG-AUUUGUUUAUUUUCCACGCGU-CA-AAAGGAAAAUAAAUGCGGAA-UCGAAAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACUGGUUCACGGGUGGAA ...-...........((((((.(((-..-................((((-(((((.((....(((((....)))))..)))))).))))).....(((....)))))).)))))) ( -31.00) >DroEre_CAF1 58273 111 + 1 AGG-AUCUGUUUAUUUUCCACGCGA-CG-AAAGGAAAAUAAAUGCGGAA-UCGACAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACCGGUUCUUAGGUGGAA .((-.((((((((((((((......-..-...)))))))))))..((((-((((.(((....(((((....)))))..)))))).)))))....))).))..((((.....)))) ( -31.80) >DroMoj_CAF1 93517 106 + 1 GGCUAUUUAUUUAUUUUCCACGGCAAAA-AAAGGAAAAUAAAAGAGGAA-UCGCUAGCAUAAACACCACGCGGUGUUCGCAACAAAUUCCAAAAAGAGCUGAUGU------AGA- ((((.(((.((((((((((.........-...))))))))))...((((-(.....((...((((((....)))))).)).....)))))..))).)))).....------...- ( -24.80) >DroAna_CAF1 38501 112 + 1 AGG-AUUUGUUUAUUUUCCACGCGA-CA-AAAGGAAAAUAAAACGGGAAUUCGACAGCAUAAACACCACGCGGUGUUCGCUCCGUAUUCCAAAAAGAGCCGGUUCAAGGGUGGAA ...-...........((((((.(..-.(-(..((...........(((((.((..(((...((((((....)))))).))).)).)))))........))..))...).)))))) ( -27.31) >consensus AGG_AUUUGUUUAUUUUCCACGCGA_CA_AAAGGAAAAUAAAUGCGGAA_UCGAAAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACCGGUUC__GGGUGGAA ........(((((((((((.............)))))))))))..((((.....((((....(((((....)))))..))))....))))......................... (-20.30 = -20.60 + 0.31)
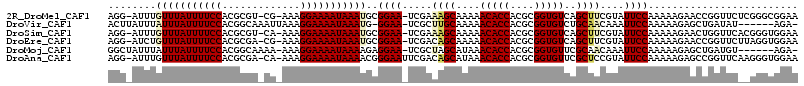
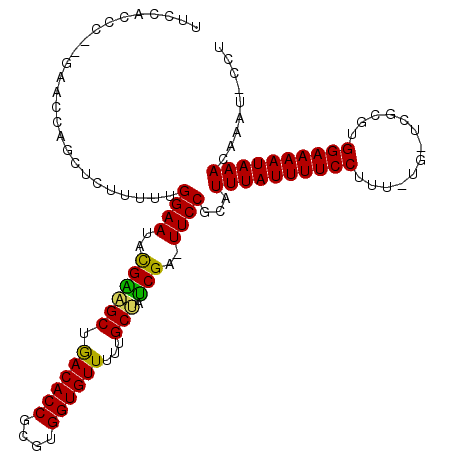
| Location | 4,910,450 – 4,910,561 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -24.00 |
| Energy contribution | -22.97 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
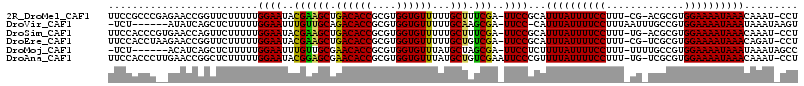
>2R_DroMel_CAF1 4910450 111 - 20766785 UUCCGCCCGAGAACCGGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUUUCGA-UUCCGCAUUUAUUUUCCUUU-CG-ACGCGUGGAAAAUAAACAAAU-CCU ....(((((.....)))........(((((.((((((.((((((....))))))...)).)))))-)))))).((((((((((...-..-......)))))))))).....-... ( -32.70) >DroVir_CAF1 75533 106 - 1 -UCU------AUAUCAGCUCUUUUUGGAAUUUGUUGCAGACACCGCGUGGUGUUUUUGCAAGCGA-UUCC-CAUUUAUUUUCCUUUAAUUUGCCGUGGAAAAUAAAUAAAUAAGU -...------......(((..(((.((((((..(((((((((((....))))))..)))))..))-))))-.(((((((((((.............))))))))))))))..))) ( -28.02) >DroSim_CAF1 62290 111 - 1 UUCCACCCGUGAACCAGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUUUCGA-UUCCGCAUUUAUUUUCCUUU-UG-ACGCGUGGAAAAUAAACAAAU-CCU ((((((.((((((....))).....(((((.((((((.((((((....))))))...)).)))))-))))................-..-))).))))))...........-... ( -31.70) >DroEre_CAF1 58273 111 - 1 UUCCACCUAAGAACCGGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUGUCGA-UUCCGCAUUUAUUUUCCUUU-CG-UCGCGUGGAAAAUAAACAGAU-CCU ...............((.(((....(((((.((((((.((((((....))))))...))).))))-))))...((((((((((...-..-......)))))))))).))).-)). ( -31.90) >DroMoj_CAF1 93517 106 - 1 -UCU------ACAUCAGCUCUUUUUGGAAUUUGUUGCGAACACCGCGUGGUGUUUAUGCUAGCGA-UUCCUCUUUUAUUUUCCUUU-UUUUGCCGUGGAAAAUAAAUAAAUAGCC -...------......(((.(((..((((((.((((((((((((....)))))))..)).)))))-))))...((((((((((...-.........)))))))))).))).))). ( -26.80) >DroAna_CAF1 38501 112 - 1 UUCCACCCUUGAACCGGCUCUUUUUGGAAUACGGAGCGAACACCGCGUGGUGUUUAUGCUGUCGAAUUCCCGUUUUAUUUUCCUUU-UG-UCGCGUGGAAAAUAAACAAAU-CCU ...............((........(((((.(((((((((((((....)))))))..))).))).))))).((((.(((((((...-..-......)))))))))))....-)). ( -27.60) >consensus UUCCACCC__GAACCAGCUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUAUCGA_UUCCGCAUUUAUUUUCCUUU_UG_UCGCGUGGAAAAUAAACAAAU_CCU .........................((((..((((((.((((((....))))))...))).)))..))))...((((((((((.............))))))))))......... (-24.00 = -22.97 + -1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:15 2006