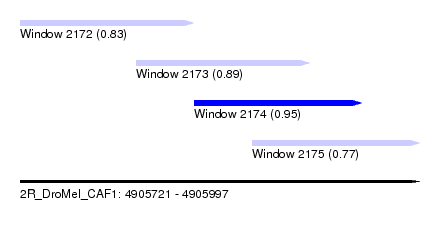
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,905,721 – 4,905,997 |
| Length | 276 |
| Max. P | 0.951207 |

| Location | 4,905,721 – 4,905,841 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -32.94 |
| Energy contribution | -31.74 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4905721 120 + 20766785 AACCACCAUCCGAGCCUUGGAUGCCCAGCGCGUUCUGGAGAAGGAGUUCGACAGCUACCAGGAUGCGCACAGCUCUGCAUUUUUCAUGUACAUAAGUACAUCGCAGGCAUUUGGUUAUUG (((((.(((((((...)))))))....(((((((((((((..(........)..)).)))))))))))...((.((((.......((((((....)))))).))))))...))))).... ( -42.50) >DroSec_CAF1 38960 120 + 1 AACCACCAUCCGAGCCCAGGAUGCCCAGCGCGUUCUGGAGAAGGAGUUCGAUAGCUACCAGGAUGCGCACAGCUCCGCAUUUUUCAUGUACAUAAGUACCUCGAAGGCAUUUGGUUAUAG (((((((...((((....(((.((...(((((((((((......(((......))).)))))))))))...)))))...........((((....))))))))..))....))))).... ( -36.10) >DroSim_CAF1 57565 120 + 1 AACCACCAUCCGAGCCCUGGAUGCCCAGAGCGUUCUGGAGAAGGAGUUCGACAGCUACCAGGAUGCGCACAGCUCUGCAUUUUUCAUGUACAUAAGUACCUCGAAGGCAUUUGGUUAUUG (((((......((((.((((....)))).(((((((((((..(........)..)).))))))))).....))))(((....(((..((((....))))...))).)))..))))).... ( -35.80) >DroEre_CAF1 53535 120 + 1 AACAACCAUCCGAGCCUUGGAGGCUCAGCGCGUUCUGGAGAAGGAGUUCGAUAAUUACCAGGAUGCGCACAGCUCGGCAUUCUACAUGUACAUAAGUACCUCGGUGGCUUUUGGUUAUUG ...(((((...(((((.(.((((((..(((((((((((...................)))))))))))..)))..............((((....))))))).).))))).))))).... ( -39.81) >DroYak_CAF1 58159 120 + 1 AACAACCAUCCGAGCCUUGGAAGCUCAGCGCGUUCUGGAGAAGGAGUUCGACAAUUACCAGGAUGCGCACAGCUCUGCAUUUUACAUGUACAUAAGUACUUCGAUGGCUUUCGGUUAUUA ...((((....((((((((((((((..(((((((((((.((..(.......)..)).)))))))))))..)))).............((((....))))))))).)))))..)))).... ( -37.30) >consensus AACCACCAUCCGAGCCUUGGAUGCCCAGCGCGUUCUGGAGAAGGAGUUCGACAGCUACCAGGAUGCGCACAGCUCUGCAUUUUUCAUGUACAUAAGUACCUCGAAGGCAUUUGGUUAUUG ............((((..(((((((..(((((((((((((..(........)..)).)))))))))))...................((((....))))......))))))))))).... (-32.94 = -31.74 + -1.20)

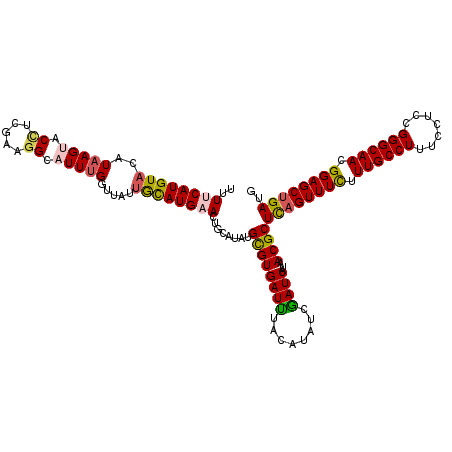
| Location | 4,905,801 – 4,905,921 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4905801 120 + 20766785 UUUUCAUGUACAUAAGUACAUCGCAGGCAUUUGGUUAUUGUAUGAACUGCAUAUGCGUGAUUUAUAUAUCGAUCAUAACGCUUAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUG .....((((((....)))))).....((...(((((..((((((((((((....))).).))))))))..)))))....))((((((((.((((((.......)))))).)))))))).. ( -32.30) >DroSec_CAF1 39040 120 + 1 UUUUCAUGUACAUAAGUACCUCGAAGGCAUUUGGUUAUAGCAUGAACUGCAUAUGUGUGAUCUACAUAUCGAUCAUAACGCUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUG ..(((((((((.(((((.((.....)).)))))))....)))))))..((.....(((((((........)))))))..))((((((((.((((((.......)))))).)))))))).. ( -34.50) >DroSim_CAF1 57645 120 + 1 UUUUCAUGUACAUAAGUACCUCGAAGGCAUUUGGUUAUUGCAUGAACUGCAUAUGUGUGAUCUACAUAUCGAUCAUAACGCUCAGUUUUUUUGCCUUUCCUCCGGGCAACGGAGCGGAUG ..((((((((..(((((.((.....)).))))).....)))))))).........(((((((........))))))).(((((.((.....(((((.......))))))).))))).... ( -31.20) >DroEre_CAF1 53615 120 + 1 UCUACAUGUACAUAAGUACCUCGGUGGCUUUUGGUUAUUGCAUGAACUGCAUAUGCGUGAUUUACAUAUCAAUCAUCACGCUCAGUUUCUUUGCCUUUCCGCCGGGCAACGGAGCUGAUG ....((((((((.((((((....)).)))).)).....))))))..........(((((((.............)))))))((((((((.((((((.......)))))).)))))))).. ( -34.42) >DroYak_CAF1 58239 120 + 1 UUUACAUGUACAUAAGUACUUCGAUGGCUUUCGGUUAUUAUAUGAACAUCAUAUGCGUGAUUUACAUAUCGAUCAUUACGCUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUG .......((((....))))...((((((.....))))))(((((.....)))))(((((((.............)))))))((((((((.((((((.......)))))).)))))))).. ( -31.92) >consensus UUUUCAUGUACAUAAGUACCUCGAAGGCAUUUGGUUAUUGCAUGAACUGCAUAUGCGUGAUUUACAUAUCGAUCAUAACGCUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUG ..((((((((..(((((.((.....)).))))).....))))))))........((((((((........))))...))))((((((((.((((((.......)))))).)))))))).. (-26.06 = -26.18 + 0.12)

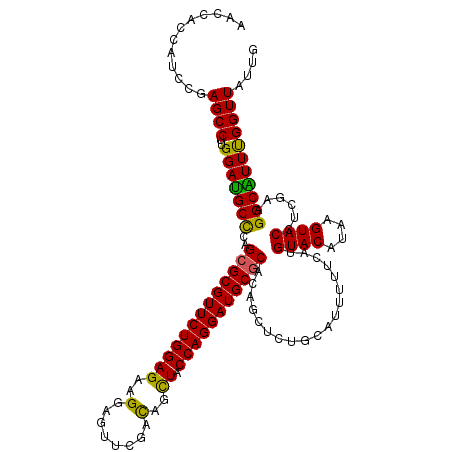
| Location | 4,905,841 – 4,905,957 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.07 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -28.14 |
| Energy contribution | -26.82 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
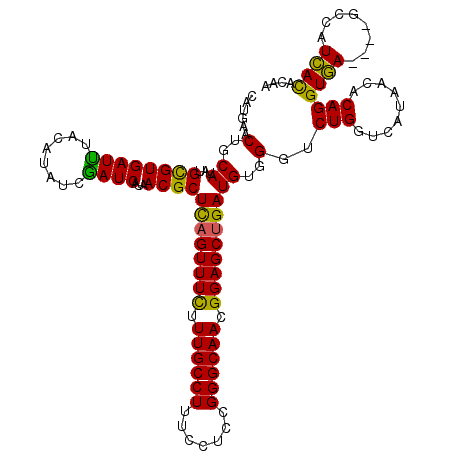
>2R_DroMel_CAF1 4905841 116 + 20766785 UAUGAACUGCAUAUGCGUGAUUUAUAUAUCGAUCAUAACGCUUAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUGUUGGUCUGGUCAUAACACAGGUAA----GCCAUUACACUA ........((....))(((((.........((((((((((.((((((((.((((((.......)))))).)))))))))))))...)))))........((...----.))))))).... ( -29.50) >DroSec_CAF1 39080 120 + 1 CAUGAACUGCAUAUGUGUGAUCUACAUAUCGAUCAUAACGCUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUGUGGGUCUGCUCAUAACACAGGUGAGCGAGCCAUCACACAA .............((((((((.................(((((((((((.((((((.......)))))).)))))))).)))((..(((((((.......)))))))..)))))))))). ( -45.20) >DroSim_CAF1 57685 120 + 1 CAUGAACUGCAUAUGUGUGAUCUACAUAUCGAUCAUAACGCUCAGUUUUUUUGCCUUUCCUCCGGGCAACGGAGCGGAUGUGGGUCUGGUCAUAACACAGGUGAGCGAGCCAUCACACAA .............((((((((...((((((((.(.....).)).((((..((((((.......))))))..)))).))))))((..((.((((.......)))).))..)))))))))). ( -37.00) >DroEre_CAF1 53655 116 + 1 CAUGAACUGCAUAUGCGUGAUUUACAUAUCAAUCAUCACGCUCAGUUUCUUUGCCUUUCCGCCGGGCAACGGAGCUGAUGUGGGUCUGGUCAUAACACAGGUGA----GCCAUCAUACAA .((((.(..((...(((((((.............)))))))((((((((.((((((.......)))))).))))))))))..)...(((((((.......))))----.))))))).... ( -36.52) >DroYak_CAF1 58279 115 + 1 UAUGAACAUCAUAUGCGUGAUUUACAUAUCGAUCAUUACGCUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUGUCGGUCUGGUCAUCACACAGGUGA----G-CAUCAUACAA (((((...((((.((.(((((...((.(((((.((......((((((((.((((((.......)))))).))))))))))))))).))...))))).)).))))----.-..)))))... ( -38.10) >consensus CAUGAACUGCAUAUGCGUGAUUUACAUAUCGAUCAUAACGCUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUGUGGGUCUGGUCAUAACACAGGUGA____GCCAUCACACAA ......(..((...((((((((........))))...))))((((((((.((((((.......)))))).))))))))))..)..(((.........)))((((........)))).... (-28.14 = -26.82 + -1.32)


| Location | 4,905,881 – 4,905,997 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.24 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.63 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
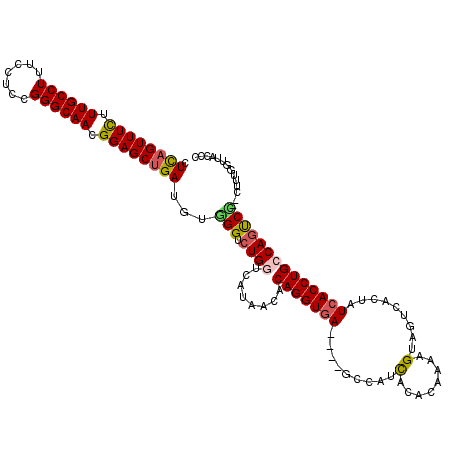
>2R_DroMel_CAF1 4905881 116 + 20766785 CUUAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUGUUGGUCUGGUCAUAACACAGGUAA----GCCAUUACACUAAAGCAGUCACUAUCACCUGCCAAUCACCUUUUGGUUACCC .((((((((.((((((.......)))))).)))))))).((((((..(((.(((.....((...----.)).....(((.....)))...))).))).)))))).(((....)))..... ( -27.80) >DroSec_CAF1 39120 118 + 1 CUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUGUGGGUCUGCUCAUAACACAGGUGAGCGAGCCAUCACACAAAUGUAGUCACUAUCACCUGUCAGUCU--CUUUGGUUACCC .((((((((.((((((.......)))))).))))))))((((((....))))))..((((((((...((..((.(((....))).))..)).))))))))......--............ ( -36.00) >DroSim_CAF1 57725 118 + 1 CUCAGUUUUUUUGCCUUUCCUCCGGGCAACGGAGCGGAUGUGGGUCUGGUCAUAACACAGGUGAGCGAGCCAUCACACAAAAGUAGUCACUAUCACCUGACAGUCC--CUUUGGUUACCC .((.((((..((((((.......))))))..)))).))...(((.(((((....)).(((((((...((..((.((......)).))..)).))))))).))).))--)........... ( -32.40) >DroEre_CAF1 53695 114 + 1 CUCAGUUUCUUUGCCUUUCCGCCGGGCAACGGAGCUGAUGUGGGUCUGGUCAUAACACAGGUGA----GCCAUCAUACAAAGGUAUUCACCAUCACCUGCCAGUCC--CUUUGAUUCUCC .((((((((.((((((.......)))))).))))))))...(((.((((........(((((((----......((((....))))......))))))))))).))--)........... ( -40.50) >DroYak_CAF1 58319 113 + 1 CUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUGUCGGUCUGGUCAUCACACAGGUGA----G-CAUCAUACAAAAGUAUUCACCAUCACCUGCCAGCCC--UUUUGGUUCCCC .((((((((.((((((.......)))))).))))))))....((.((((........(((((((----.-....((((....))))......))))))))))).))--............ ( -36.10) >consensus CUCAGUUUCUUUGCCUUUCCUCCGGGCAACGGAGCUGAUGUGGGUCUGGUCAUAACACAGGUGA____GCCAUCACACAAAAGUAGUCACUAUCACCUGCCAGUCC__CUUUGGUUACCC .((((((((.((((((.......)))))).))))))))...(((.((((........(((((((.........(........).........)))))))))))))).............. (-25.39 = -25.63 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:10 2006