
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,882,116 – 4,882,219 |
| Length | 103 |
| Max. P | 0.896936 |

| Location | 4,882,116 – 4,882,219 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.31 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -18.96 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
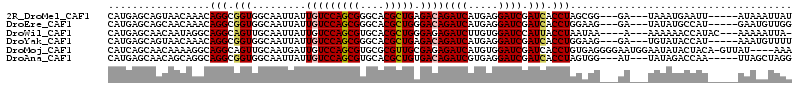
>2R_DroMel_CAF1 4882116 103 + 20766785 CAUGAGCAGUAACAAACAGGCGGUGGCAAUUAUUGUCCAGCGGGCACGCUGAGACAGAUCAUGAGGAUCGAUCACCUAGCGG---GA---UAAAUGAAUU-----AUAAAUUAU .....((.((.....))....((((((......(((((((((....))))).))))((((.....))))).)))))..))..---..---..........-----......... ( -25.40) >DroEre_CAF1 29653 103 + 1 CAUGAGCAGCAACAAACAGGCGGUGGCAAUUAUUGUCCAGCGGGCACGCUGGGACAGAUCAUGAGGAUCGAUCACCUGGAAG---GA---UAUAUGCCAU-----GAAUGUUGG ((((.(((........((((.(((.........(((((((((....)))).)))))((((.....)))).))).))))....---..---....))))))-----)........ ( -32.67) >DroWil_CAF1 23179 103 + 1 CAUGAGCAACAAUAGGCAGGCAGUUGCAAUUAUUGUCCAGCGUGCACGCUGGGAGAGAUCUUGUGGAUCCAUUACCUAAUAA----A---AAAAAACCAUAC---AAAAAUUA- .(((.(((((............)))))..(((((((((((((....))))))).(.((((.....)))))......))))))----.---.......)))..---........- ( -22.30) >DroYak_CAF1 34077 103 + 1 CAUGAGCAGUAACAAACAGGCGGUGGCAAUUAUUGUCCAGCGGGCACGCUGAGACAGAUCAUGAGGAUCGAUCACCUGGAAG---GA---UGUAUACCAU-----AAAUGUUUU ...(((((........((((.(((.........(((((((((....))))).))))((((.....)))).))).))))...(---(.---......))..-----...))))). ( -28.40) >DroMoj_CAF1 63267 109 + 1 CAUCAGCAACAAAAGGCAGGCAGUUGCAAUGAUUGUCCAGCGUGCGCGUUGCGAGAGAUCAUGUGGAUCGAUCACCUGUGAGGGGAAUGGAAUAUACUACA-GUUAU----AAA .....(((((............))))).((((((.(((((((....))))).))..))))))((((.((.((..(((....)))..)).)).....)))).-.....----... ( -25.30) >DroAna_CAF1 15767 103 + 1 CAUGAGCAACAGCAGGCAGGCGGUGGCAAUUAUUGUCCAGCGUGCACGCUGUGACAGAUCGUGAGGAUCGAUCACCUAGUGG---AU---UAUAGACCAA-----UUAGCUAGG .....(..(((((..(((.((.(.((((.....))))).)).)))..)))))..).(((((.......))))).((((((.(---((---(.......))-----)).)))))) ( -33.70) >consensus CAUGAGCAACAACAAACAGGCGGUGGCAAUUAUUGUCCAGCGGGCACGCUGAGACAGAUCAUGAGGAUCGAUCACCUAGAAG___GA___UAUAUACCAU_____AAAAAUUAG .................(((.(((.........(((((((((....))))).))))((((.....)))).))).)))..................................... (-18.96 = -19.10 + 0.14)


| Location | 4,882,116 – 4,882,219 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.31 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.07 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
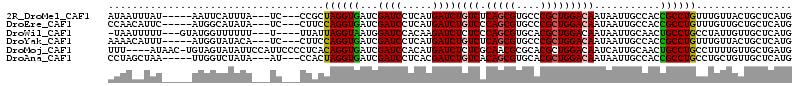
>2R_DroMel_CAF1 4882116 103 - 20766785 AUAAUUUAU-----AAUUCAUUUA---UC---CCGCUAGGUGAUCGAUCCUCAUGAUCUGUCUCAGCGUGCCCGCUGGACAAUAAUUGCCACCGCCUGUUUGUUACUGCUCAUG .........-----..........---..---....((((((...((((.....))))((((.(((((....)))))))))...........))))))................ ( -21.90) >DroEre_CAF1 29653 103 - 1 CCAACAUUC-----AUGGCAUAUA---UC---CUUCCAGGUGAUCGAUCCUCAUGAUCUGUCCCAGCGUGCCCGCUGGACAAUAAUUGCCACCGCCUGUUUGUUGCUGCUCAUG .(((((..(-----(.(((.....---..---......((((...((((.....))))(((((.((((....)))))))))........)))))))))..)))))......... ( -28.91) >DroWil_CAF1 23179 103 - 1 -UAAUUUUU---GUAUGGUUUUUU---U----UUAUUAGGUAAUGGAUCCACAAGAUCUCUCCCAGCGUGCACGCUGGACAAUAAUUGCAACUGCCUGCCUAUUGUUGCUCAUG -.((((.((---((..........---.----............(((((.....)))))...((((((....)))))))))).))))(((((............)))))..... ( -22.30) >DroYak_CAF1 34077 103 - 1 AAAACAUUU-----AUGGUAUACA---UC---CUUCCAGGUGAUCGAUCCUCAUGAUCUGUCUCAGCGUGCCCGCUGGACAAUAAUUGCCACCGCCUGUUUGUUACUGCUCAUG ........(-----((((((.(((---..---....((((((...((((.....))))((((.(((((....)))))))))...........))))))..)))...))).)))) ( -26.30) >DroMoj_CAF1 63267 109 - 1 UUU----AUAAC-UGUAGUAUAUUCCAUUCCCCUCACAGGUGAUCGAUCCACAUGAUCUCUCGCAACGCGCACGCUGGACAAUCAUUGCAACUGCCUGCCUUUUGUUGCUGAUG ...----.....-..........((((............((((..((((.....))))..))))...((....))))))..((((..(((((............))))))))). ( -18.90) >DroAna_CAF1 15767 103 - 1 CCUAGCUAA-----UUGGUCUAUA---AU---CCACUAGGUGAUCGAUCCUCACGAUCUGUCACAGCGUGCACGCUGGACAAUAAUUGCCACCGCCUGCCUGCUGUUGCUCAUG (((((....-----.(((......---..---)))))))).(((((.......))))).((.((((((.(((.(((((.((.....)))))..)).))).)))))).))..... ( -29.30) >consensus AUAACAUAU_____AUGGUAUAUA___UC___CUACCAGGUGAUCGAUCCUCAUGAUCUGUCCCAGCGUGCACGCUGGACAAUAAUUGCCACCGCCUGCCUGUUGCUGCUCAUG ....................................((((((...((((.....))))((((.(((((....)))))))))...........))))))................ (-20.18 = -20.07 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:58 2006