| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,881,350 – 4,881,441 |
| Length | 91 |
| Max. P | 0.996491 |

| Location | 4,881,350 – 4,881,441 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.64 |
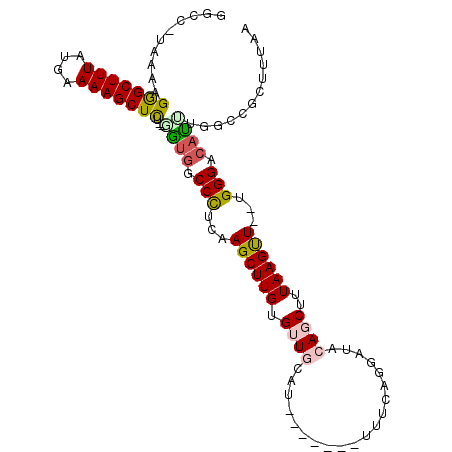
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -16.50 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
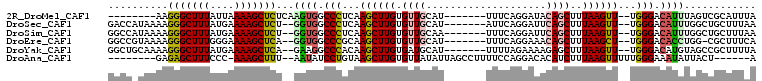
>2R_DroMel_CAF1 4881350 91 - 20766785 --------AAGGGCUUUAUUAAAAGCUCUCAAGUGGCCCUCAAGCUUGUGUUGCAU-------UUUCAGGAUACAGCUUUAAGUU--UGGGACAUUUAGUCGCAUUUA --------.((((((((....))))))))...(((((.((((((((((.((((.((-------(.....))).))))..))))))--)))).......)))))..... ( -25.50) >DroSec_CAF1 14590 97 - 1 GACCAUAAAAGGGCUUUAUGAAAAGCUCU--GGUGGCCCUCAAGCUUGUGUUGCAU-------AUUCAGGAUUCAGCUUUAAGUU--UGGGACAUUUGGCUGCUUUAA .........((((((((....))))))))--((..(((((((((((((.((((.((-------.......)).))))..))))))--))))......)))..)).... ( -28.60) >DroSim_CAF1 33235 97 - 1 GGCCAUAAAAGGGCUUUAUGAAAAGCUCU--GGUGGCCCUCAAGCUUGUGUUGCAA-------UUUCAGGAUUCAGCUUUAAGUU--UGGGACAUUUGGCUGCUUUAA ((((((...((((((((....))))))))--.))))))((((((((((.((((.((-------((....))))))))..))))))--))))................. ( -35.50) >DroEre_CAF1 28905 96 - 1 GGCCGUAAAAGGGCUUUGGGAAAAGCUCA--GGUGGCCCGCAAGCUUGUGUUGCAU-------UUUCAGGAAACAGCUUUAAGCU--UGGGACACCUGG-CGCUUUCA .(((......(((((((....)))))))(--((((..((.((((((((.((((..(-------(.....))..))))..))))))--)))).)))))))-)....... ( -36.60) >DroYak_CAF1 33273 97 - 1 GGCUGCAAAAGGGCUUUAUGAAAAGCUCA--GAAGGCCCACAAGCUUGUGAUGCAU-------UUUUAGAAAAGAGCUUUAAGUU--UGGGACAUGUAGCCGCUUUUA (((((((...(((((((.(((.....)))--.))))))).((((((((....((.(-------(((....)))).))..))))))--)).....)))))))....... ( -34.30) >DroAna_CAF1 14958 91 - 1 --------GAGAGCUUUCCC-AAAGCUUU--AAUAUCCUGUAAGCUUGUGUUAUAUUAGCCUUUUCCAGGACACAUCUUUAAGUUUUUGGGAAAUAUUACU------A --------......((((((-((((.(((--((..(((((.(((((.(((...))).)).)))...))))).......))))).)))))))))).......------. ( -18.30) >consensus GGCC_UAAAAGGGCUUUAUGAAAAGCUCU__GGUGGCCCUCAAGCUUGUGUUGCAU_______UUUCAGGAUACAGCUUUAAGUU__UGGGACAUUUGGCCGCUUUAA ..........(((((((....)))))))...((((.(((...((((((.((((....................))))..))))))...))).))))............ (-16.50 = -16.75 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:56 2006