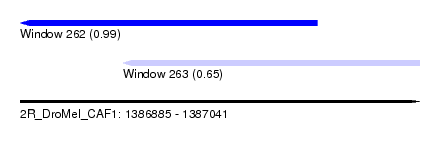
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,386,885 – 1,387,041 |
| Length | 156 |
| Max. P | 0.987166 |

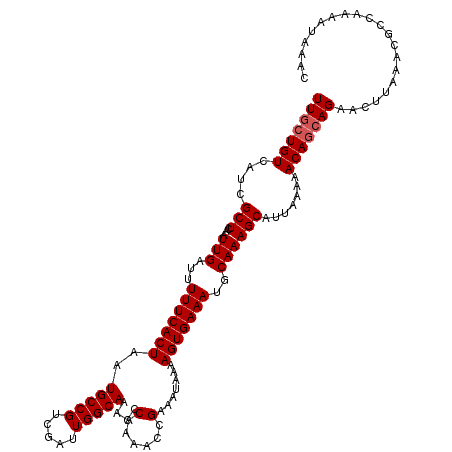
| Location | 1,386,885 – 1,387,001 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 97.93 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -18.78 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1386885 116 - 20766785 UUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAACCCAAAACCGAAAUAAAAGUGAAAUGCAAAGCAUUAAAAACAGCAGAACUUAAACGCCAAAAUAAAC (((((((....(((...(((...(((((((..(((((.....)))))....(......).......)))))))..)))))).......)))))))..................... ( -21.40) >DroSec_CAF1 13977 116 - 1 UUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAAACCAAAACCGAAAUAAAAGUGAAAUGCAAAGCAUUAAAAACAGCAGAACUUAAACGCCAAAAUAAAC (((((((....(((...(((...(((((((..(((((.....)))))....(......).......)))))))..)))))).......)))))))..................... ( -21.40) >DroSim_CAF1 18713 116 - 1 UUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAACCCAAAACCGAAAUAAAAGUGAAAUGCAAAGCAUUAAAAACAGCAGAACUUAAACGCCAAAAUAAAC (((((((....(((...(((...(((((((..(((((.....)))))....(......).......)))))))..)))))).......)))))))..................... ( -21.40) >DroEre_CAF1 12099 116 - 1 UUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAACCCAAAACCGAAAUAAAAGUGAAAUGCAAAGCAUUAAAAACAGAAGAACUUAAACGCCAAAAUAAAC ...((((....(((...(((...(((((((..(((((.....)))))....(......).......)))))))..)))))).......))))........................ ( -16.30) >DroYak_CAF1 9899 116 - 1 UUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCAAUUGGCAAACGCAAAACCGAAAUAAAAGUGAAAUGCAAAGCAUUAAAAACAACAGAACUUAAACGCCAAAAUAAAU ...........(((...(((...(((((((..(((((.....)))))..((......)).......)))))))..))))))................................... ( -17.20) >consensus UUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAACCCAAAACCGAAAUAAAAGUGAAAUGCAAAGCAUUAAAAACAGCAGAACUUAAACGCCAAAAUAAAC (((((((....(((...(((...(((((((..(((((.....)))))....(......).......)))))))..)))))).......)))))))..................... (-18.78 = -19.18 + 0.40)

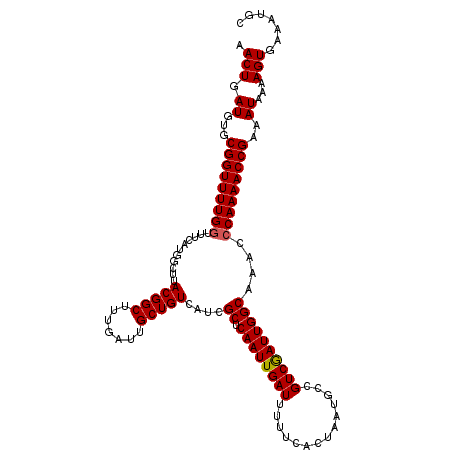
| Location | 1,386,925 – 1,387,041 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 98.62 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -28.36 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1386925 116 - 20766785 AACUGAUGUGCGGUUUUGGUUUCAUGGCUUACGGCUUUGAUUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAACCCAAAACCGAAAUAAAAGUGAAAUGC .(((.((...(((((((((..(((.(((..(((((.......)))))....)))....)))...........(((((.....)))))...)))))))))..))...)))....... ( -29.60) >DroSec_CAF1 14017 116 - 1 AACUGAUGUGCGGUUUUGGUUUCAUGGCUUACGGCUUUGAUUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAAACCAAAACCGAAAUAAAAGUGAAAUGC .(((.((...((((((((((((........(((((.......)))))....((.((((((((..............)))))))))).))))))))))))..))...)))....... ( -32.24) >DroSim_CAF1 18753 116 - 1 AACUGAUGUGCGGUUUUGGUUUCAUGGCUUACGGCUUUGAUUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAACCCAAAACCGAAAUAAAAGUGAAAUGC .(((.((...(((((((((..(((.(((..(((((.......)))))....)))....)))...........(((((.....)))))...)))))))))..))...)))....... ( -29.60) >DroEre_CAF1 12139 116 - 1 AACUGAUGUGCGGUUUUGGUUUCAUGGCUUACGGCUUUGAUUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAACCCAAAACCGAAAUAAAAGUGAAAUGC .(((.((...(((((((((..(((.(((..(((((.......)))))....)))....)))...........(((((.....)))))...)))))))))..))...)))....... ( -29.60) >DroYak_CAF1 9939 116 - 1 AACUGAUGUGCGGUUUUGGUUUUAUGGCUUACGGCUUUGAUUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCAAUUGGCAAACGCAAAACCGAAAUAAAAGUGAAAUGC .(((.((...((((((((((((........(((((.......)))))....((.((((((((..............)))))))))))))).))))))))..))...)))....... ( -28.74) >consensus AACUGAUGUGCGGUUUUGGUUUCAUGGCUUACGGCUUUGAUUGCUGUCAUCGCUCAAUUGAUUUUUCACUAAUGCCGUCGAUUGGCAAACCCAAAACCGAAAUAAAAGUGAAAUGC .(((.((...(((((((((...........(((((.......)))))....((.((((((((..............))))))))))....)))))))))..))...)))....... (-28.36 = -28.40 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:28:01 2006