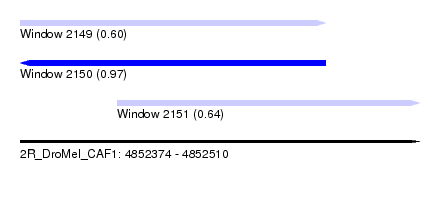
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,852,374 – 4,852,510 |
| Length | 136 |
| Max. P | 0.969443 |

| Location | 4,852,374 – 4,852,478 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -29.67 |
| Energy contribution | -30.01 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
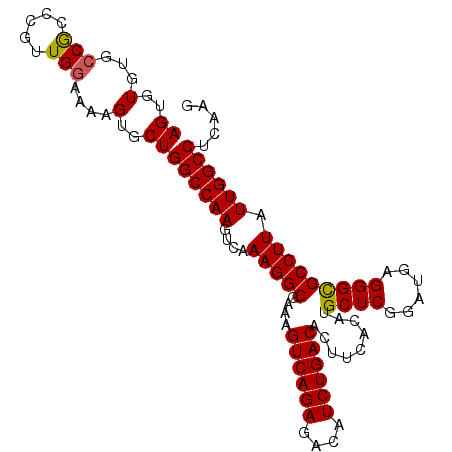
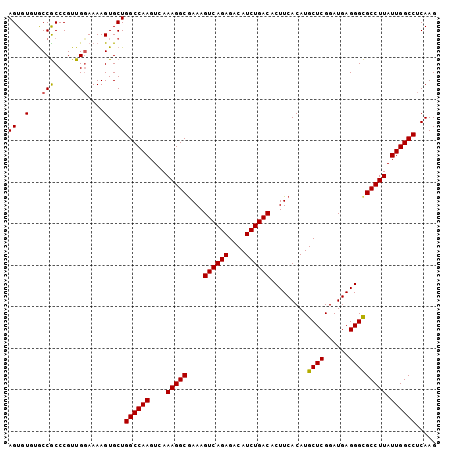
>2R_DroMel_CAF1 4852374 104 + 20766785 GCAGAUGAGUGGCCAUAUGAAUGUGGGCGUAAGAGUGUGCACCGCCCGUUGGAAAAGUGCUGGCCAAGUCAAAGGCAAAAGUCAGAGACAUCUGACACUUCACA ((...(((.(((((((((.....((((((.............))))))........))).))))))..)))...))....((((((....))))))........ ( -31.94) >DroEre_CAF1 8761 104 + 1 GCAGAUGAGUGGCCAUAUGAAUGUGGGCGUAAGAGUGUUUGCCGCCCGUUGGAAAAGUGCUGGCCAAGUCAAAGGCGAAAGUCAGAGACAUCUGACACUUCACA ((...(((.(((((((((.....((((((((((....)))).))))))........))).))))))..)))...))(((.((((((....))))))..)))... ( -34.02) >DroYak_CAF1 13207 104 + 1 GCAGAUGAGUGGCCAUAUGAAUGUGGGCGUAAGAGUGUGUGCCACCCGUUGCAAAAGUGCUGGCCAAGUCAAAGGCGAAAGUCAGAGACAUCUGACACUUCACA (((.(((.((((((((((..(((....)))....))))).))))).))))))....(((..((((........)))....((((((....)))))).)..))). ( -35.30) >consensus GCAGAUGAGUGGCCAUAUGAAUGUGGGCGUAAGAGUGUGUGCCGCCCGUUGGAAAAGUGCUGGCCAAGUCAAAGGCGAAAGUCAGAGACAUCUGACACUUCACA ((...(((.(((((((((.....((((((.............))))))........))).))))))..)))...))....((((((....))))))........ (-29.67 = -30.01 + 0.33)


| Location | 4,852,374 – 4,852,478 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -30.56 |
| Energy contribution | -30.12 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4852374 104 - 20766785 UGUGAAGUGUCAGAUGUCUCUGACUUUUGCCUUUGACUUGGCCAGCACUUUUCCAACGGGCGGUGCACACUCUUACGCCCACAUUCAUAUGGCCACUCAUCUGC .(..(((.((((((....)))))))))..)...(((..((((((.............(((((.............))))).........)))))).)))..... ( -28.77) >DroEre_CAF1 8761 104 - 1 UGUGAAGUGUCAGAUGUCUCUGACUUUCGCCUUUGACUUGGCCAGCACUUUUCCAACGGGCGGCAAACACUCUUACGCCCACAUUCAUAUGGCCACUCAUCUGC .((((((.((((((....))))))))))))...(((..((((((.............(((((.............))))).........)))))).)))..... ( -31.17) >DroYak_CAF1 13207 104 - 1 UGUGAAGUGUCAGAUGUCUCUGACUUUCGCCUUUGACUUGGCCAGCACUUUUGCAACGGGUGGCACACACUCUUACGCCCACAUUCAUAUGGCCACUCAUCUGC .((((((.((((((....))))))))))))...(((..(((((((((....)))...(((((.............))))).........)))))).)))..... ( -34.22) >consensus UGUGAAGUGUCAGAUGUCUCUGACUUUCGCCUUUGACUUGGCCAGCACUUUUCCAACGGGCGGCACACACUCUUACGCCCACAUUCAUAUGGCCACUCAUCUGC .((((((.((((((....))))))))))))...(((..(((((((........)...(((((.............))))).........)))))).)))..... (-30.56 = -30.12 + -0.44)

| Location | 4,852,407 – 4,852,510 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 95.47 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -32.71 |
| Energy contribution | -32.60 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4852407 103 + 20766785 AGUGUGCACCGCCCGUUGGAAAAGUGCUGGCCAAGUCAAAGGCAAAAGUCAGAGACAUCUGACACUUCACAUGCUCGGAUGAGGGCGCCUUAUUGGCCUCAAG .....((((..((....))....)))).((((((....(((((....((((((....)))))).........((((......))))))))).))))))..... ( -37.30) >DroEre_CAF1 8794 103 + 1 AGUGUUUGCCGCCCGUUGGAAAAGUGCUGGCCAAGUCAAAGGCGAAAGUCAGAGACAUCUGACACUUCACAUGCUCGGAUGAGGGCGCCUUAUUGGCCUCAAG ((..(((....((....))..)))..))((((((....((((((((.((((((....))))))..)))....((((......))))))))).))))))..... ( -36.50) >DroYak_CAF1 13240 103 + 1 AGUGUGUGCCACCCGUUGCAAAAGUGCUGGCCAAGUCAAAGGCGAAAGUCAGAGACAUCUGACACUUCACAUGCUCGGAUGAGGGUGCCUUAUUGGCCUCAAG ((..(.(((........)))...)..))((((((....((((((((.((((((....))))))..)))....((((......))))))))).))))))..... ( -34.30) >consensus AGUGUGUGCCGCCCGUUGGAAAAGUGCUGGCCAAGUCAAAGGCGAAAGUCAGAGACAUCUGACACUUCACAUGCUCGGAUGAGGGCGCCUUAUUGGCCUCAAG ((..(...(((.....)))....)..))((((((....(((((....((((((....)))))).........((((......))))))))).))))))..... (-32.71 = -32.60 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:47 2006