| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,851,295 – 4,851,495 |
| Length | 200 |
| Max. P | 0.972630 |

| Location | 4,851,295 – 4,851,415 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -28.21 |
| Energy contribution | -28.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4851295 120 + 20766785 UUAAAAUCAAUUAGAGAUUCGGUCGAAACAUGCACUCAGGUAAACGAUGCUUAACUGCCAGGCUAUCUAAUCUGAUUUUAAUUAAGCUGAUAGUCUGCUAUUUGCAGGCGCCCUGCAAUC (((((((((((((((.....((((.......(((.(.(((((.....))))).).)))..)))).)))))).)))))))))....((.....((((((.....)))))).....)).... ( -29.10) >DroEre_CAF1 7779 120 + 1 UUAAAAUCAAUUAGAGAUUCGCUCGCAACAUGCACUCAGGUAAACGAUGCUUAACUGACAGGGUAUCUAAUCUGAUUUUAAUUAAGCUGAUAGUCUGCUCUGUGCAGGCGCACUGCAAUC (((((((((((((((.....((((((.....))..((((.(((.......))).))))..)))).)))))).)))))))))....((.....((((((.....)))))).....)).... ( -31.20) >DroYak_CAF1 12155 120 + 1 UUAAAAUCAAUUAGAGAUUCGGUCGAAACGUGCACUCAGGUAAACGAUGCUUAACUGCCAGGGUAUCUAAUCUGAUUUUAAUUAAGCUGAUAGUCUGCUCUUUGCAGGCGCACUGCAAUC (((((((((((((((.((((.((((..((.((....)).))...))))((......))..)))).)))))).)))))))))....((.....((((((.....)))))).....)).... ( -31.90) >consensus UUAAAAUCAAUUAGAGAUUCGGUCGAAACAUGCACUCAGGUAAACGAUGCUUAACUGCCAGGGUAUCUAAUCUGAUUUUAAUUAAGCUGAUAGUCUGCUCUUUGCAGGCGCACUGCAAUC (((((((((((((((.(((((((........(((.((........)))))......))).)))).)))))).)))))))))....((.....((((((.....)))))).....)).... (-28.21 = -28.87 + 0.67)

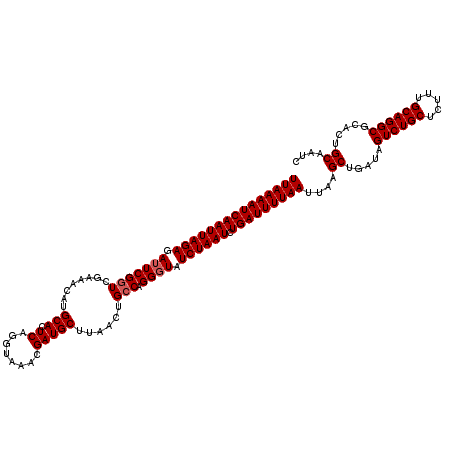
| Location | 4,851,375 – 4,851,495 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4851375 120 + 20766785 AUUAAGCUGAUAGUCUGCUAUUUGCAGGCGCCCUGCAAUCCGGCACUGGAAAAAACUCGGUAUCCUUUAUAAACAUACCUAUCACAUAACAAAUAAAUUAACGCAGCAAAAUAAAUAUUA .....((((.((((..(((..(((((((...)))))))...)))))))..........(((((...........)))))........................))))............. ( -22.40) >DroEre_CAF1 7859 104 + 1 AUUAAGCUGAUAGUCUGCUCUGUGCAGGCGCACUGCAAUCCGGCACUGGAAAAAACUGUGCAGCCU-CAUGGAGAUG--CCUCACGUAACAAGGAAUGGAAUAUAA----A--------- .....((.....((((((.....)))))).....))..((((((((.(.......).))))..(((-.((((((...--.))).)))....)))..))))......----.--------- ( -26.50) >DroYak_CAF1 12235 117 + 1 AUUAAGCUGAUAGUCUGCUCUUUGCAGGCGCACUGCAAUCCGGCACUGCAAAAAUCUAUGCAUAAA-UGUAUGUACG--UAUCACAUAUCAAAUAAUUCAAUACAACAAAAUAUAUAUUA .......(((((((((((.....))))))(((.(((......))).))).........((((((..-..))))))..--.......)))))............................. ( -23.10) >consensus AUUAAGCUGAUAGUCUGCUCUUUGCAGGCGCACUGCAAUCCGGCACUGGAAAAAACUAUGCAUCCU_UAUAAACAUG__UAUCACAUAACAAAUAAUUCAAUACAACAAAAUA_AUAUUA .....((((...((((((.....))))))((...))....))))............................................................................ (-15.20 = -15.20 + 0.00)

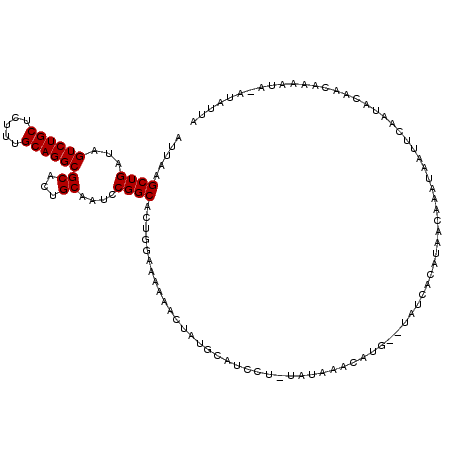
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:44 2006