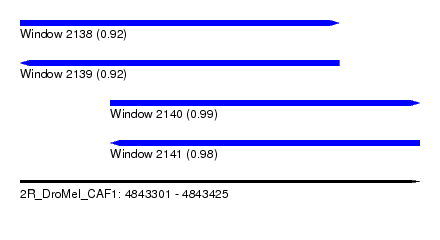
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,843,301 – 4,843,425 |
| Length | 124 |
| Max. P | 0.991533 |

| Location | 4,843,301 – 4,843,400 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -28.23 |
| Energy contribution | -27.57 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4843301 99 + 20766785 GGAGCUGGCUUAUUACUUGCAAUGAAUAAAUAAACAUGGGAUCGUGCGAACGCACAAAUGUACCACACACAGGA---------UGUGCCUGUGUUCAUGUUUAUGCGA ...((..((.........)).........((((((((((....((((....))))...........(((((((.---------....))))))))))))))))))).. ( -27.90) >DroSim_CAF1 3915 99 + 1 GGAGCUGGCUUAUUACUUGCAAUGAAUAAAUAAACAUGGGAUCGUGCGAACGCACAAAUGUACUACACACAGGA---------UGUGCCUGUGCCCAUGUUUAUGCGA ...((..((.........)).........(((((((((((...((((....))))............((((((.---------....))))))))))))))))))).. ( -32.60) >DroYak_CAF1 4226 108 + 1 GAAGCUGGCUUAUUACUUGCAAUGAAUAAAUAAACAUGGGAUCGUGCGAACGCACAAAUGUACUACACACAGGGUUCUGCGAGUGUGUCUGUGUCCAUGUUUAUGCCC ......((((((((......)))))....(((((((((((...((((....)))).....(((.((((((..(......)..))))))..))))))))))))))))). ( -29.90) >consensus GGAGCUGGCUUAUUACUUGCAAUGAAUAAAUAAACAUGGGAUCGUGCGAACGCACAAAUGUACUACACACAGGA_________UGUGCCUGUGUCCAUGUUUAUGCGA ...((..((.........)).........(((((((((((...((((....))))............((((((..............))))))))))))))))))).. (-28.23 = -27.57 + -0.66)

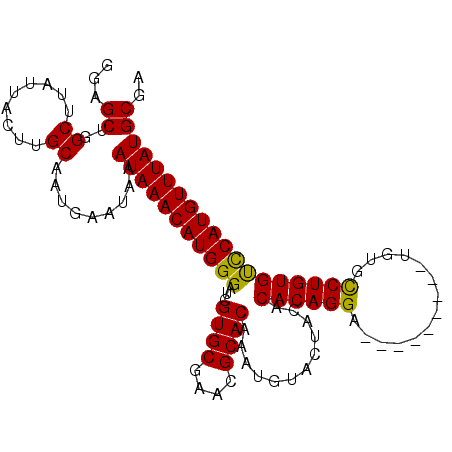
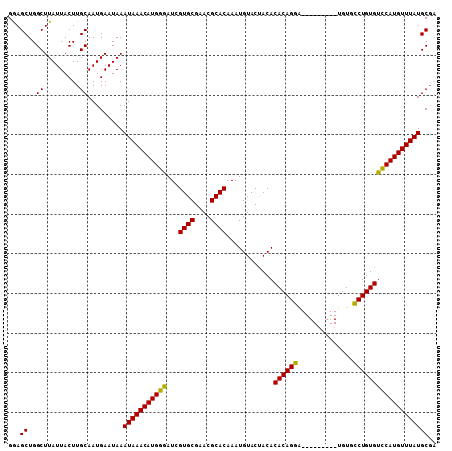
| Location | 4,843,301 – 4,843,400 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4843301 99 - 20766785 UCGCAUAAACAUGAACACAGGCACA---------UCCUGUGUGUGGUACAUUUGUGCGUUCGCACGAUCCCAUGUUUAUUUAUUCAUUGCAAGUAAUAAGCCAGCUCC ..(((((((((((.(((((((....---------.)))))))..((.....((((((....)))))).)))))))))))......((((....))))......))... ( -27.00) >DroSim_CAF1 3915 99 - 1 UCGCAUAAACAUGGGCACAGGCACA---------UCCUGUGUGUAGUACAUUUGUGCGUUCGCACGAUCCCAUGUUUAUUUAUUCAUUGCAAGUAAUAAGCCAGCUCC ..(((((((((((((((((((....---------.))))))).........((((((....))))))..))))))))))......((((....))))......))... ( -30.40) >DroYak_CAF1 4226 108 - 1 GGGCAUAAACAUGGACACAGACACACUCGCAGAACCCUGUGUGUAGUACAUUUGUGCGUUCGCACGAUCCCAUGUUUAUUUAUUCAUUGCAAGUAAUAAGCCAGCUUC .(((((((((((((........((((.(((((....))))).)).))....((((((....))))))..))))))))))......((((....))))..)))...... ( -31.40) >consensus UCGCAUAAACAUGGACACAGGCACA_________UCCUGUGUGUAGUACAUUUGUGCGUUCGCACGAUCCCAUGUUUAUUUAUUCAUUGCAAGUAAUAAGCCAGCUCC ..((((((((((((((((((................)))))).........((((((....))))))..))))))))))......((((....))))......))... (-25.31 = -25.42 + 0.11)

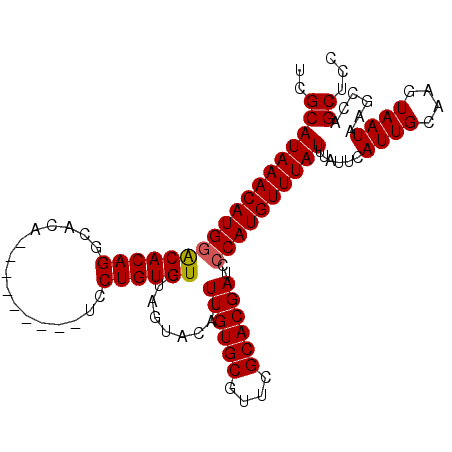
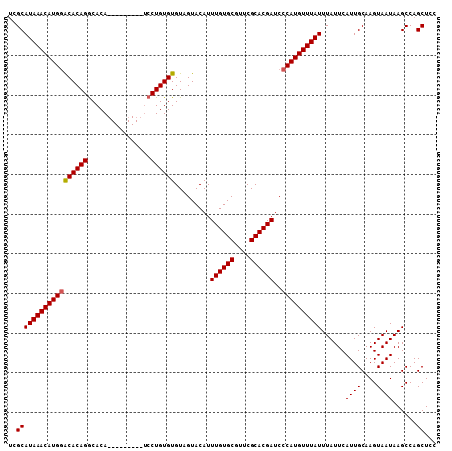
| Location | 4,843,329 – 4,843,425 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -28.20 |
| Energy contribution | -27.87 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4843329 96 + 20766785 AAUAAACAUGGGAUCGUGCGAACGCACAAAUGUACCACACACAGGA---------UGUGCCUGUGUUCAUGUUUAUGCGAUGCCCAUUUCAUGAAAUGAAUUCCU--------------- .((((((((((....((((....))))...........(((((((.---------....)))))))))))))))))........(((((....))))).......--------------- ( -27.30) >DroSim_CAF1 3943 111 + 1 AAUAAACAUGGGAUCGUGCGAACGCACAAAUGUACUACACACAGGA---------UGUGCCUGUGCCCAUGUUUAUGCGAUGCGCAUUUCACGAAAUGAAUUCCUUGUGUGCAAACACGC .(((((((((((...((((....))))............((((((.---------....)))))))))))))))))(((.(((((((.....(((.....)))...)))))))....))) ( -39.40) >DroYak_CAF1 4254 120 + 1 AAUAAACAUGGGAUCGUGCGAACGCACAAAUGUACUACACACAGGGUUCUGCGAGUGUGUCUGUGUCCAUGUUUAUGCCCUGCCCAUUUCACGAAAUGAAUUCCCGGUGUGCAAACACAG .(((((((((((...((((....)))).....(((.((((((..(......)..))))))..))))))))))))))........(((((....)))))........((((....)))).. ( -32.20) >consensus AAUAAACAUGGGAUCGUGCGAACGCACAAAUGUACUACACACAGGA_________UGUGCCUGUGUCCAUGUUUAUGCGAUGCCCAUUUCACGAAAUGAAUUCCU_GUGUGCAAACAC__ .(((((((((((...((((....))))............((((((..............)))))))))))))))))........(((((....)))))........((((....)))).. (-28.20 = -27.87 + -0.33)

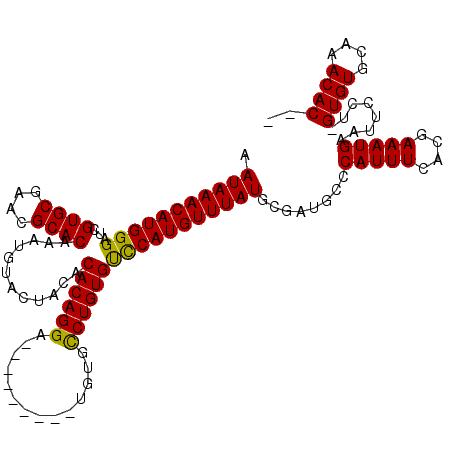
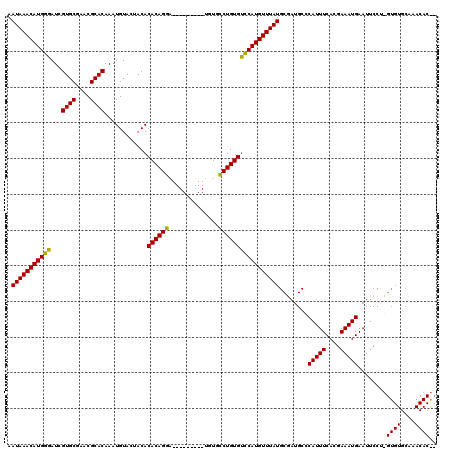
| Location | 4,843,329 – 4,843,425 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -25.71 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4843329 96 - 20766785 ---------------AGGAAUUCAUUUCAUGAAAUGGGCAUCGCAUAAACAUGAACACAGGCACA---------UCCUGUGUGUGGUACAUUUGUGCGUUCGCACGAUCCCAUGUUUAUU ---------------.((..(((((((....)))))))..))..(((((((((.(((((((....---------.)))))))..((.....((((((....)))))).))))))))))). ( -28.90) >DroSim_CAF1 3943 111 - 1 GCGUGUUUGCACACAAGGAAUUCAUUUCGUGAAAUGCGCAUCGCAUAAACAUGGGCACAGGCACA---------UCCUGUGUGUAGUACAUUUGUGCGUUCGCACGAUCCCAUGUUUAUU ((((((((.(((.....(....).....))))))))))).....(((((((((((((((((....---------.))))))).........((((((....))))))..)))))))))). ( -39.70) >DroYak_CAF1 4254 120 - 1 CUGUGUUUGCACACCGGGAAUUCAUUUCGUGAAAUGGGCAGGGCAUAAACAUGGACACAGACACACUCGCAGAACCCUGUGUGUAGUACAUUUGUGCGUUCGCACGAUCCCAUGUUUAUU .((((....))))((..(...((((((....)))))).).))..((((((((((........((((.(((((....))))).)).))....((((((....))))))..)))))))))). ( -33.90) >consensus __GUGUUUGCACAC_AGGAAUUCAUUUCGUGAAAUGGGCAUCGCAUAAACAUGGACACAGGCACA_________UCCUGUGUGUAGUACAUUUGUGCGUUCGCACGAUCCCAUGUUUAUU ....................(((((((....)))))))......((((((((((((((((................)))))).........((((((....))))))..)))))))))). (-25.71 = -26.16 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:38 2006