
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,841,464 – 4,841,559 |
| Length | 95 |
| Max. P | 0.999831 |

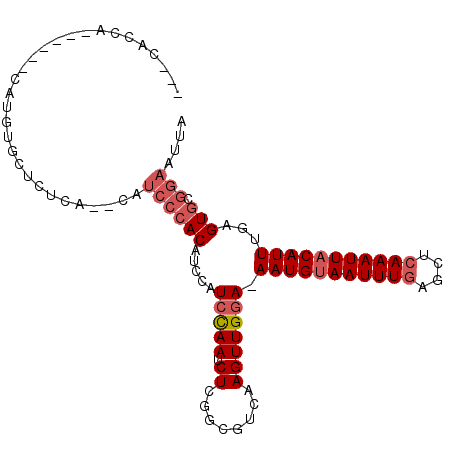
| Location | 4,841,464 – 4,841,559 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.75 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -13.24 |
| Energy contribution | -14.80 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
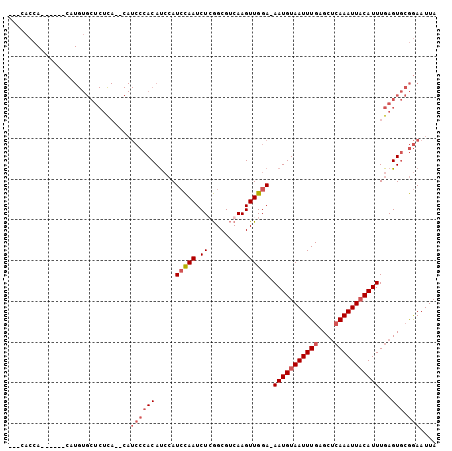
>2R_DroMel_CAF1 4841464 95 + 20766785 ---CACCA------CAUGUGCUCUCACACAUCCCACAUCCAUCCAAUCUGGGCGUCAAGUUGGA-AAUGUAAUUUGAGCUCAAAUUACAUUUGAGUGCGGAAUUA ---.....------..((((......))))((((((.....(((((..((.....))..)))))-(((((((((((....)))))))))))...))).))).... ( -26.60) >DroVir_CAF1 11193 73 + 1 ---CA-----------------------------ACACAAAUGUAAUCUGAGCGGCAAGUUGGAAAAUGUAAUUUGAGCGAAAAUUUCAUUUGAGUAGAAAAUUA ---..-----------------------------((.((((((.(((.(..((..(((((((.......))))))).))..).))).)))))).))......... ( -12.90) >DroSim_CAF1 2119 93 + 1 ---AACCA------CAUGUGCUCUCA--CAUCCCACAUCCAUCCAAUCUCGGCGUCAAGUUGGA-AAUGUAAUUUGAGCUCAAAUUACAUUUGAGUGCGGAAUUA ---.....------..((((....))--))((((((............(((((.....)))))(-(((((((((((....))))))))))))..))).))).... ( -23.50) >DroYak_CAF1 2468 102 + 1 AAGCACCACAUGCCCAUGUGCUCUUA--CAUCCCACAUCCAUCCAAUCUCGGCGUCAAGUUGGA-AAUGUAAUUUGAGCUCAAAUUACAUUUGAGUGCGGAAUUA (((...(((((....)))))..))).--..((((((............(((((.....)))))(-(((((((((((....))))))))))))..))).))).... ( -25.80) >consensus ___CACCA______CAUGUGCUCUCA__CAUCCCACAUCCAUCCAAUCUCGGCGUCAAGUUGGA_AAUGUAAUUUGAGCUCAAAUUACAUUUGAGUGCGGAAUUA ..............................((((((.....(((((.((........))))))).(((((((((((....)))))))))))...))).))).... (-13.24 = -14.80 + 1.56)

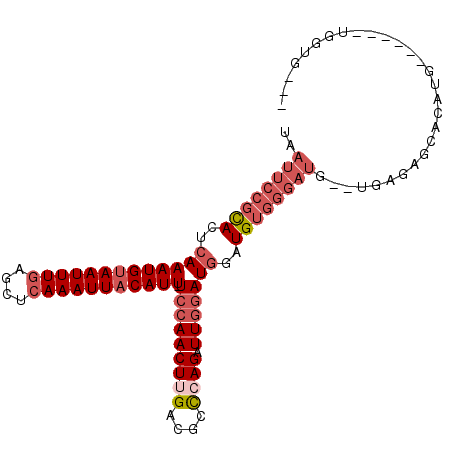
| Location | 4,841,464 – 4,841,559 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 74.75 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -17.15 |
| Energy contribution | -20.03 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.61 |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
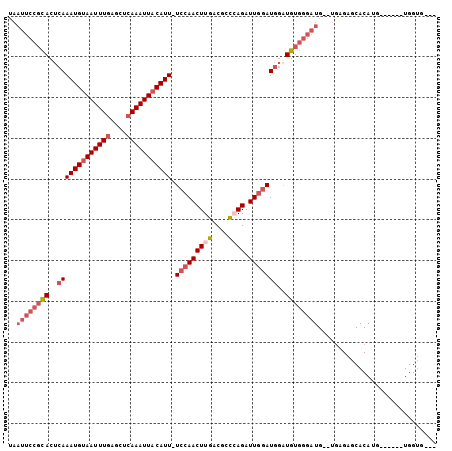
>2R_DroMel_CAF1 4841464 95 - 20766785 UAAUUCCGCACUCAAAUGUAAUUUGAGCUCAAAUUACAUU-UCCAACUUGACGCCCAGAUUGGAUGGAUGUGGGAUGUGUGAGAGCACAUG------UGGUG--- ..((((((((..(((((((((((((....)))))))))))-(((((.(((.....))).)))))))..))))))))((((....))))...------.....--- ( -34.70) >DroVir_CAF1 11193 73 - 1 UAAUUUUCUACUCAAAUGAAAUUUUCGCUCAAAUUACAUUUUCCAACUUGCCGCUCAGAUUACAUUUGUGU-----------------------------UG--- .........((.((((((.(((((..((.(((...............)))..))..))))).)))))).))-----------------------------..--- ( -7.86) >DroSim_CAF1 2119 93 - 1 UAAUUCCGCACUCAAAUGUAAUUUGAGCUCAAAUUACAUU-UCCAACUUGACGCCGAGAUUGGAUGGAUGUGGGAUG--UGAGAGCACAUG------UGGUU--- ..((((((((..(((((((((((((....)))))))))))-(((((((((....)))).)))))))..))))))))(--(....)).....------.....--- ( -32.60) >DroYak_CAF1 2468 102 - 1 UAAUUCCGCACUCAAAUGUAAUUUGAGCUCAAAUUACAUU-UCCAACUUGACGCCGAGAUUGGAUGGAUGUGGGAUG--UAAGAGCACAUGGGCAUGUGGUGCUU ..((((((((..(((((((((((((....)))))))))))-(((((((((....)))).)))))))..)))))))).--...((((((((......)).)))))) ( -36.60) >consensus UAAUUCCGCACUCAAAUGUAAUUUGAGCUCAAAUUACAUU_UCCAACUUGACGCCCAGAUUGGAUGGAUGUGGGAUG__UGAGAGCACAUG______UGGUG___ ..((((((((..(((((((((((((....))))))))))).(((((((((....)))).)))))))..))))))))............................. (-17.15 = -20.03 + 2.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:34 2006