
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,840,340 – 4,840,500 |
| Length | 160 |
| Max. P | 0.981009 |

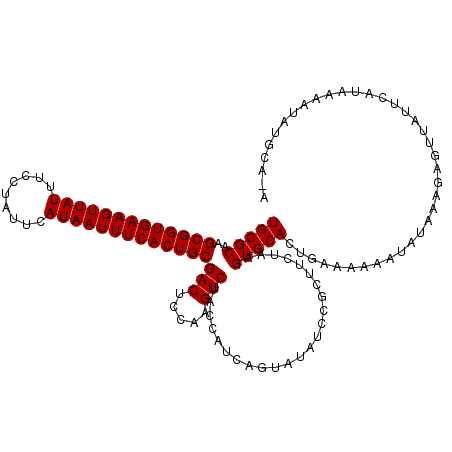
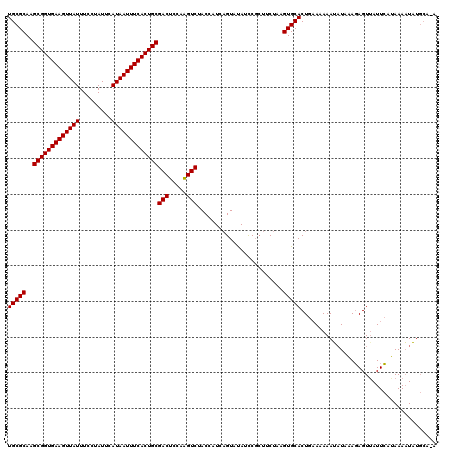
| Location | 4,840,340 – 4,840,460 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4840340 120 - 20766785 UGCGCAAGCGGUGAAGUUAUUUCCUAUUCAUAAUUUCACUGCGACUCCAAGUCUACCAUCAGUUUAUCCGCUUCUAAGUGCACUGAAAAAAUAUAAAGAGUAAUUCAUAAAAUAUGCAAA ...(((.(((((((((((((.........))))))))))))).((((...........(((((.....((((....)))).)))))...........)))).............)))... ( -26.95) >DroSim_CAF1 994 120 - 1 UGCGCAAGCGGUGAAGUUAUUUCCUAUUCAUAAUUUCACUGCGACUCCAAGUCGACCAUCAGUACAUCCGCUUCUAAGUGCACUGAAAAAAUAUUAAGAGUUAUUCAUAAAAUAUGCACA .(.(((.(((((((((((((.........)))))))))))))(((((..(((......(((((.(((..........))).)))))......)))..)))))............))).). ( -28.40) >DroYak_CAF1 1322 114 - 1 UGCGCAAGCGGUGAAGUUAUUUCCUAUUCAUAAUUUCACUGCGACUCCAGGUCUACCAUUUGUAUAUUCUAUUCUAAGUGCACUGUAAAAAG-UGUACAGUUAUUGCAAAAUUCU----- ...(((((((((((((((((.........)))))))))))))((((...((....))....................(((((((......))-))))))))).))))........----- ( -32.70) >consensus UGCGCAAGCGGUGAAGUUAUUUCCUAUUCAUAAUUUCACUGCGACUCCAAGUCUACCAUCAGUAUAUCCGCUUCUAAGUGCACUGAAAAAAUAUAAAGAGUUAUUCAUAAAAUAUGCA_A (((((..(((((((((((((.........)))))))))))))(((.....)))........................)))))...................................... (-24.50 = -24.50 + 0.00)

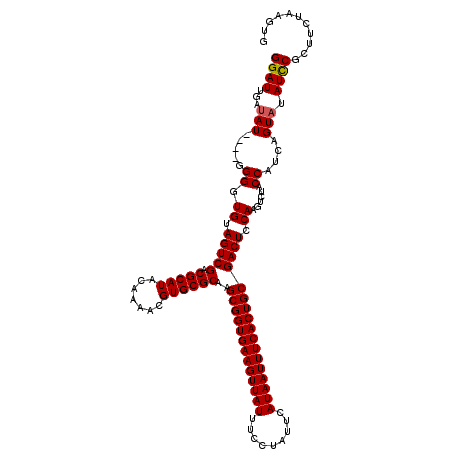
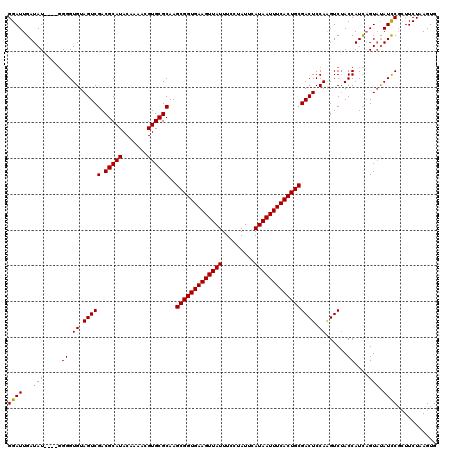
| Location | 4,840,380 – 4,840,500 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -35.43 |
| Consensus MFE | -28.95 |
| Energy contribution | -29.07 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4840380 120 - 20766785 GGAUUGAUAUAGGGGGGGUGUAGUCGACGCAUACAAAACGUGCGCAAGCGGUGAAGUUAUUUCCUAUUCAUAAUUUCACUGCGACUCCAAGUCUACCAUCAGUUUAUCCGCUUCUAAGUG ((((((((...((..((.((.(((((.(((((.......))))))..(((((((((((((.........))))))))))))))))).))..))..))))))))))...((((....)))) ( -36.80) >DroSim_CAF1 1034 116 - 1 GGAUUGAUAU----GGGGUGUAGUCGACGCAUACAAAACGUGCGCAAGCGGUGAAGUUAUUUCCUAUUCAUAAUUUCACUGCGACUCCAAGUCGACCAUCAGUACAUCCGCUUCUAAGUG ((.(((((.(----(((((......(.(((((.......))))))..(((((((((((((.........))))))))))))).)))))).)))))))...........((((....)))) ( -37.10) >DroYak_CAF1 1356 115 - 1 GGAUUGAUAU-----GGGUGUAGUCGACGCAUACAAAACGUGCGCAAGCGGUGAAGUUAUUUCCUAUUCAUAAUUUCACUGCGACUCCAGGUCUACCAUUUGUAUAUUCUAUUCUAAGUG (((...((((-----..(((.(((((.(((((.......))))))..(((((((((((((.........))))))))))))))))).).((....))))..))))..))).......... ( -32.40) >consensus GGAUUGAUAU____GGGGUGUAGUCGACGCAUACAAAACGUGCGCAAGCGGUGAAGUUAUUUCCUAUUCAUAAUUUCACUGCGACUCCAAGUCUACCAUCAGUAUAUCCGCUUCUAAGUG ((((...(((.....((.((.(((((.(((((.......))))))..(((((((((((((.........))))))))))))))))).))......))....))).))))........... (-28.95 = -29.07 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:31 2006