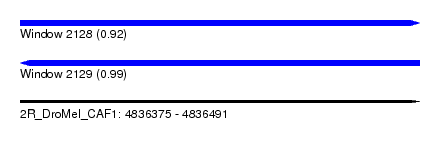
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,836,375 – 4,836,491 |
| Length | 116 |
| Max. P | 0.992033 |

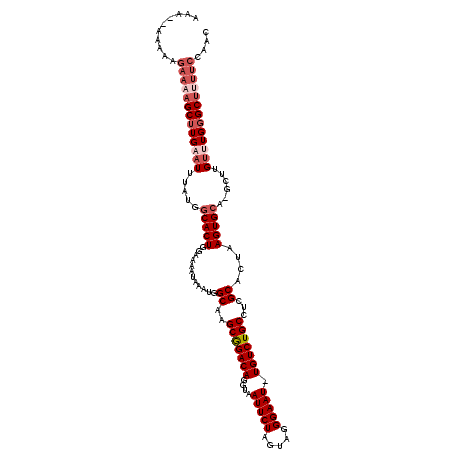
| Location | 4,836,375 – 4,836,491 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -19.84 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4836375 116 + 20766785 GUUGGAAAAGCCCAAACAAGC-UGCACUUAGUGCGAGGCAGACA-AUUCCCUACUAGAAUUACCUGUCCGCUUGCAAUUUAUUUUCCAGUGCCAUAAAUUCAAGCUUUUCUUUUU--UUU ...((((((((.....(((((-.((((...))))(.(((((..(-((((.......)))))..))))))))))).(((((((...........)))))))...))))))))....--... ( -26.80) >DroSec_CAF1 6949 104 + 1 GUUGGAAAAGCCCAAACAAGCCUGCACUUAGUGCGAGGCAGACA-AUUCCCUACUAGAAUUACCUGUCCGCUUGCCAUUUAUUUUCCAGUGCCAUAAAUUCAAGC--------------- .((((((((.......(((((..((((...))))(.(((((..(-((((.......)))))..))))))))))).......))))))))................--------------- ( -25.94) >DroSim_CAF1 9790 119 + 1 GUUGGAAAAGCCCAAACAAGCCUGCACUUAGUGCGAGGCAGACA-AUUCCCUACUAGAAUUACCUGUCCGCUUGCCAUUUAUUUUCCAGUGCCAUAAAUUCAAGCUUUUCUUUUUUUUUU ...((((((((.....(((((..((((...))))(.(((((..(-((((.......)))))..))))))))))).((((........))))............))))))))......... ( -26.10) >DroEre_CAF1 12097 116 + 1 GUCUGAAAAGCCCAAACAAGC-UGCACUUAGAGCCGGGCAGACAUAUUCCCUACCAGAAUUACCUGUCUGCUUGCCACUUAUUUUCCAGUGUCAUAAAUGCAAGCUUUUCCUUUU---UC ....((((((.......((((-((((.(((((..((((((((((.((((.......))))....)))))))))).((((........)))))).))).))).)))))...)))))---). ( -26.40) >DroYak_CAF1 8081 117 + 1 AGUUGAAGAGCCCAAACAAGC-UGCACUUAGUGCAAGGCAGACAUAUUCCCUACCAGAAUUACCUGUCUGCCUGCAACUUAUUUUGCAGUGCCAUAAAAGCAUGCUUUUCUUUCU--UUU ....(((((((........((-((((...((((((.((((((((.((((.......))))....))))))))))).))).....))))))((.......))..))))))).....--... ( -36.50) >consensus GUUGGAAAAGCCCAAACAAGC_UGCACUUAGUGCGAGGCAGACA_AUUCCCUACUAGAAUUACCUGUCCGCUUGCCAUUUAUUUUCCAGUGCCAUAAAUUCAAGCUUUUCUUUUU__UUU ....(((((((............(((((.(((((.((((.((((.((((.......))))....)))).)))))).)))........)))))...........))))))).......... (-19.84 = -20.20 + 0.36)

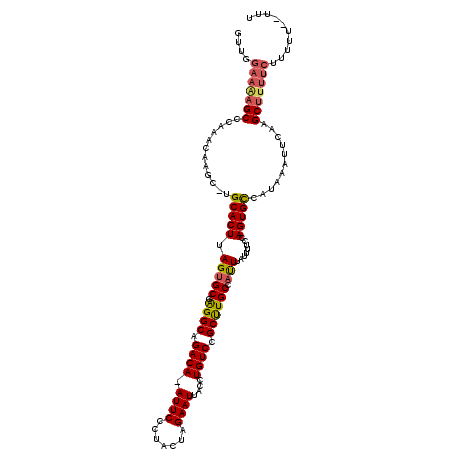
| Location | 4,836,375 – 4,836,491 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -37.24 |
| Consensus MFE | -26.79 |
| Energy contribution | -28.75 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4836375 116 - 20766785 AAA--AAAAAGAAAAGCUUGAAUUUAUGGCACUGGAAAAUAAAUUGCAAGCGGACAGGUAAUUCUAGUAGGGAAU-UGUCUGCCUCGCACUAAGUGCA-GCUUGUUUGGGCUUUUCCAAC ...--.....((((((((..(((((((..(....)...)))))).((((((((.(((..((((((.....)))))-)..)))))..((((...)))).-)))))))..)))))))).... ( -38.00) >DroSec_CAF1 6949 104 - 1 ---------------GCUUGAAUUUAUGGCACUGGAAAAUAAAUGGCAAGCGGACAGGUAAUUCUAGUAGGGAAU-UGUCUGCCUCGCACUAAGUGCAGGCUUGUUUGGGCUUUUCCAAC ---------------(((.........)))..(((((((.....(((((((((.(((..((((((.....)))))-)..)))))..((((...))))..))))))).....))))))).. ( -35.10) >DroSim_CAF1 9790 119 - 1 AAAAAAAAAAGAAAAGCUUGAAUUUAUGGCACUGGAAAAUAAAUGGCAAGCGGACAGGUAAUUCUAGUAGGGAAU-UGUCUGCCUCGCACUAAGUGCAGGCUUGUUUGGGCUUUUCCAAC ..........((((((((..(((((((..(....)...)))))).((((((((.(((..((((((.....)))))-)..)))))..((((...))))..)))))))..)))))))).... ( -38.60) >DroEre_CAF1 12097 116 - 1 GA---AAAAGGAAAAGCUUGCAUUUAUGACACUGGAAAAUAAGUGGCAAGCAGACAGGUAAUUCUGGUAGGGAAUAUGUCUGCCCGGCUCUAAGUGCA-GCUUGUUUGGGCUUUUCAGAC ..---.....((((((((((((((((.((((((........))))((..(((((((....(((((.....))))).)))))))...))))))))))))-.........)))))))).... ( -35.90) >DroYak_CAF1 8081 117 - 1 AAA--AGAAAGAAAAGCAUGCUUUUAUGGCACUGCAAAAUAAGUUGCAGGCAGACAGGUAAUUCUGGUAGGGAAUAUGUCUGCCUUGCACUAAGUGCA-GCUUGUUUGGGCUCUUCAACU ...--.....(((.(((.((((.....)))).....((((((((((((((((((((....(((((.....))))).))))))))).((.....)))))-))))))))..))).))).... ( -38.60) >consensus AAA__AAAAAGAAAAGCUUGAAUUUAUGGCACUGGAAAAUAAAUGGCAAGCGGACAGGUAAUUCUAGUAGGGAAU_UGUCUGCCUCGCACUAAGUGCA_GCUUGUUUGGGCUUUUCCAAC ..........(((((((((((((.....(((((............((..(((((((....(((((.....))))).)))))))...))....)))))......))))))))))))).... (-26.79 = -28.75 + 1.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:27 2006