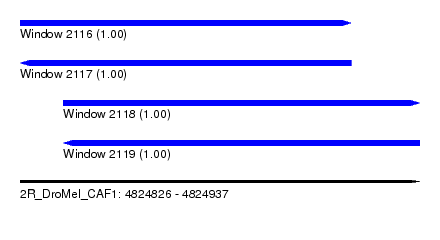
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,824,826 – 4,824,937 |
| Length | 111 |
| Max. P | 0.999357 |

| Location | 4,824,826 – 4,824,918 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -21.31 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.99 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4824826 92 + 20766785 ----------------------------UGUUGCAUUUUGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUGCAACUUUCAUCAAUCCAGACUAUUUACUUUAAAC ----------------------------..(((((.(((....))).)))))((((((((((((.(((((((((((...............))))))))..)))..)))))))))))).. ( -22.26) >DroEre_CAF1 358 92 + 1 ----------------------------UGUUGCAUUUCGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUGCAACUUUCAUCAAUCCAGACUAUUUACUUUAAAC ----------------------------..(((((.((......)).)))))((((((((((((.(((((((((((...............))))))))..)))..)))))))))))).. ( -20.56) >DroWil_CAF1 143392 120 + 1 UUUUUUUCAUUUUUUUUCUUUUCAUUUUUGUUGCAUUUUGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUAUAACUUUCAUCAAUCCAGACUAUUUACUUUAAGC ..............................(((((.(((....))).)))))((((((((((((.(((((((((((...............))))))))..)))..)))))))))))).. ( -22.26) >DroPer_CAF1 288 92 + 1 ----------------------------AGUUGCAUAUCGUUUAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUAUAACUUUCAUCAAUCCAGACUAUUUACUUUAAGC ----------------------------..(((((............)))))((((((((((((.(((((((((((...............))))))))..)))..)))))))))))).. ( -20.16) >consensus ____________________________UGUUGCAUUUCGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUACAACUUUCAUCAAUCCAGACUAUUUACUUUAAAC ..............................(((((.(((....))).)))))((((((((((((.(((((((((((...............))))))))..)))..)))))))))))).. (-20.24 = -20.99 + 0.75)

| Location | 4,824,826 – 4,824,918 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -21.75 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4824826 92 - 20766785 GUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUGCAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACAAAAUGCAACA---------------------------- (((..((((((((.(((((...(((((((..((.......))..))))))).))))))))))))).)))((((.(((....))).))))...---------------------------- ( -22.50) >DroEre_CAF1 358 92 - 1 GUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUGCAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACGAAAUGCAACA---------------------------- (((..((((((((.(((((...(((((((..((.......))..))))))).))))))))))))).)))((((.(((....))).))))...---------------------------- ( -22.90) >DroWil_CAF1 143392 120 - 1 GCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUAUAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACAAAAUGCAACAAAAAUGAAAAGAAAAAAAAUGAAAAAAA .(((.((((((((.(((((...(((((((..((.......))..))))))).)))))))))))))....((((.(((....))).))))...........)))................. ( -22.10) >DroPer_CAF1 288 92 - 1 GCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUAUAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUAAACGAUAUGCAACU---------------------------- .....((((((((.(((((...(((((((..((.......))..))))))).)))))))))))))....((((............))))...---------------------------- ( -20.60) >consensus GCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUACAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACAAAAUGCAACA____________________________ .....((((((((.(((((...(((((((..((.......))..))))))).)))))))))))))....((((.(((....))).))))............................... (-21.75 = -21.75 + 0.00)


| Location | 4,824,838 – 4,824,937 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.71 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
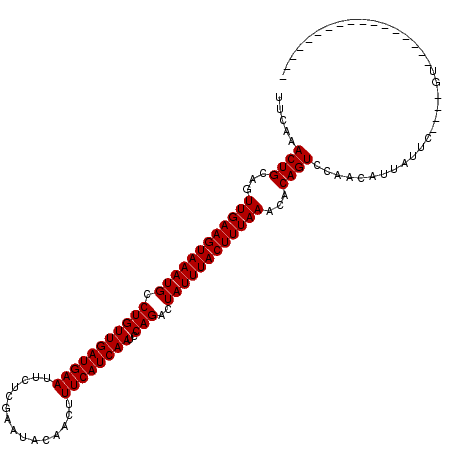
>2R_DroMel_CAF1 4824838 99 + 20766785 UUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUGCAACUUUCAUCAAUCCAGACUAUUUACUUUAAACACAGUCCAACAUUGUUC----GU----------------- .....((((...((((((((((((.(((((((((((...............))))))))..)))..))))))))))))...))))............----..----------------- ( -22.26) >DroEre_CAF1 370 99 + 1 UUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUGCAACUUUCAUCAAUCCAGACUAUUUACUUUAAACACAGUCCAACAUUGUUC----GU----------------- .....((((...((((((((((((.(((((((((((...............))))))))..)))..))))))))))))...))))............----..----------------- ( -22.26) >DroWil_CAF1 143432 120 + 1 UUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUAUAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUUCAACAUUAUGAUCUCUCCUUCUCUCUCUAUGUAU ....(((((...((((((((((((.(((((((((((...............))))))))..)))..))))))))))))...)))))..((((...((............))...)))).. ( -22.66) >DroPer_CAF1 300 105 + 1 UUUAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUAUAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUCCAACAUUAUUU----CU-----------GUGUGU ............((((((((((((.(((((((((((...............))))))))..)))..))))))))))))((((((.............----))-----------)))).. ( -24.28) >consensus UUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUACAACUUUCAUCAAUCCAGACUAUUUACUUUAAACACAGUCCAACAUUAUUC____GU_________________ .....((((...((((((((((((.(((((((((((...............))))))))..)))..))))))))))))...))))................................... (-21.71 = -21.71 + 0.00)

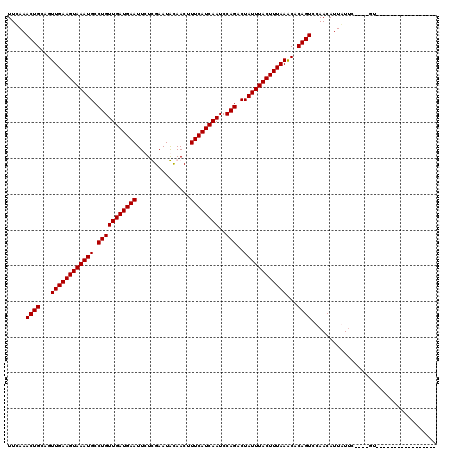
| Location | 4,824,838 – 4,824,937 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.23 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4824838 99 - 20766785 -----------------AC----GAACAAUGUUGGACUGUGUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUGCAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAA -----------------..----........(..((((((..((.((((((((.(((((...(((((((..((.......))..))))))).))))))))))))).))..))))))..). ( -29.40) >DroEre_CAF1 370 99 - 1 -----------------AC----GAACAAUGUUGGACUGUGUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUGCAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAA -----------------..----........(..((((((..((.((((((((.(((((...(((((((..((.......))..))))))).))))))))))))).))..))))))..). ( -29.40) >DroWil_CAF1 143432 120 - 1 AUACAUAGAGAGAGAAGGAGAGAUCAUAAUGUUGAACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUAUAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAA ...............................(..((((((..((.((((((((.(((((...(((((((..((.......))..))))))).))))))))))))).))..))))))..). ( -28.80) >DroPer_CAF1 300 105 - 1 ACACAC-----------AG----AAAUAAUGUUGGACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUAUAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUAAA ......-----------..----........(((((((((..((.((((((((.(((((...(((((((..((.......))..))))))).))))))))))))).))..))))))))). ( -28.70) >consensus _________________AC____GAACAAUGUUGGACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUACAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAA ...............................(((((((((..((.((((((((.(((((...(((((((..((.......))..))))))).))))))))))))).))..))))))))). (-28.60 = -28.23 + -0.38)


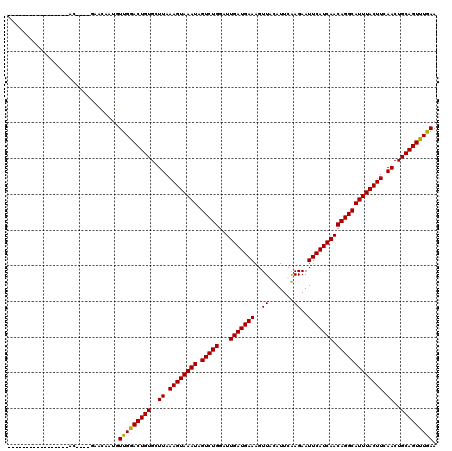
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:17 2006