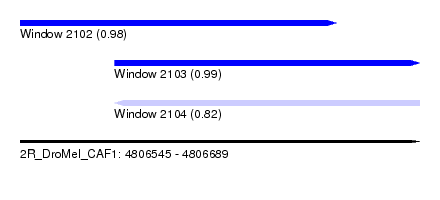
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,806,545 – 4,806,689 |
| Length | 144 |
| Max. P | 0.991165 |

| Location | 4,806,545 – 4,806,659 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -35.18 |
| Energy contribution | -36.38 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4806545 114 + 20766785 GCAGCAGCAGAAGCAGCAGCUGCUACCGUAGUGAAAUCCGAUGCAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAGGCACACAGGCAGCUAAGGACGCUG .((((.(((((.(((((.(((((....)))))....(((((((.((...)))))))))..........).)))))))))..(((((((.(.....).)))))))......)))) ( -41.60) >DroSec_CAF1 17849 108 + 1 GC------AGAAGCAGCAGCUGCUGCCGUAGUGAAAUCCGAUGCAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAGGCACACAGGCAGCUAAGGACGCUG ((------(((.(((((....)))))....((((..(((((((.((...))))))))).....)))).......)))))..(((((((.(.....).))))))).......... ( -38.30) >DroSim_CAF1 17813 114 + 1 GCAGCAGCAGAAGCAGCAGCUGCUGCCGUAGUGAAAUCCGAUGCAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAGGCACACAGGCAGCUAAGGACGCUG ((((.((((((.(((((....)))))....((((..(((((((.((...))))))))).....))))..))))))))))..(((((((.(.....).))))))).......... ( -44.10) >DroEre_CAF1 17782 114 + 1 GAAACAGCAGCAGCAGCAGCUGCUGGCGUAGUGAAAUCCGAUGGAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAAGCACACAGGCAGCAAAAGACGCUG ....((((((((((....))))))((((.((..((..(((((((......)))))))((....))....))..)).))))..((((((.........)))))).......)))) ( -44.50) >DroYak_CAF1 15508 114 + 1 GAAACAGCAGCAGCAGCAGCUGCUGCCAUAGUGAAAUCCGAUGCAGCAUCUCGUCGGGAACACUCACAGUUUGCUCUGCCAGGCUGCCAAGCACACAGGCAGCUAAAGACGCUG .((((.((((((((....))))))))....((((..(((((((.((...))))))))).....)))).))))(((((....(((((((.........)))))))..))).)).. ( -43.90) >consensus GCAACAGCAGAAGCAGCAGCUGCUGCCGUAGUGAAAUCCGAUGCAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAGGCACACAGGCAGCUAAGGACGCUG ....((((....((((.(((.((((....(((....(((((((........)))))))...)))..))))..)))))))..(((((((.........)))))))......)))) (-35.18 = -36.38 + 1.20)



| Location | 4,806,579 – 4,806,689 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -35.24 |
| Energy contribution | -35.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4806579 110 + 20766785 AAUCCGAUGCAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAGGCACACAGGCAGCUAAGGACGCUGCUUUGGCCCGUGGAAACACCACGUUCCCAC ...((((.(((((.(((.((.(((.((((.....)))).))).))..(((((((.(.....).)))))))..))).))))).))))..(((((.....)))))....... ( -39.20) >DroSec_CAF1 17877 110 + 1 AAUCCGAUGCAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAGGCACACAGGCAGCUAAGGACGCUGCUUUGGCCCGUGGAAACACCACGUUCCCAC ...((((.(((((.(((.((.(((.((((.....)))).))).))..(((((((.(.....).)))))))..))).))))).))))..(((((.....)))))....... ( -39.20) >DroSim_CAF1 17847 110 + 1 AAUCCGAUGCAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAGGCACACAGGCAGCUAAGGACGCUGCUUUGGCCCGUGGAAACACCACGUUCCCAC ...((((.(((((.(((.((.(((.((((.....)))).))).))..(((((((.(.....).)))))))..))).))))).))))..(((((.....)))))....... ( -39.20) >DroEre_CAF1 17816 110 + 1 AAUCCGAUGGAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAAGCACACAGGCAGCAAAAGACGCUGCUUUGGCCUGUGGAAAUACCAGGUUCCCAC .....(((((......)))))((((.....((.(((..(((((.....((((((.........))))))...))).)).))).))(((((.(......)))))))))).. ( -35.40) >DroYak_CAF1 15542 110 + 1 AAUCCGAUGCAGCAUCUCGUCGGGAACACUCACAGUUUGCUCUGCCAGGCUGCCAAGCACACAGGCAGCUAAAGACGCUGCUUUGGCCUGUGGAAACACCAGGUUCCCAC ...((((.(((((.(((.((.(((...((.....))...))).))..(((((((.........)))))))..))).))))).))))((((((....)).))))....... ( -38.50) >consensus AAUCCGAUGCAGCAUCUCGUCGGGAAAACUCACAGUUUGCUCUGCCAGGCUGCCAGGCACACAGGCAGCUAAGGACGCUGCUUUGGCCCGUGGAAACACCACGUUCCCAC ...((((.(((((.(((.((.(((.((((.....)))).))).))..(((((((.........)))))))..))).))))).))))...((((.(((.....))).)))) (-35.24 = -35.44 + 0.20)

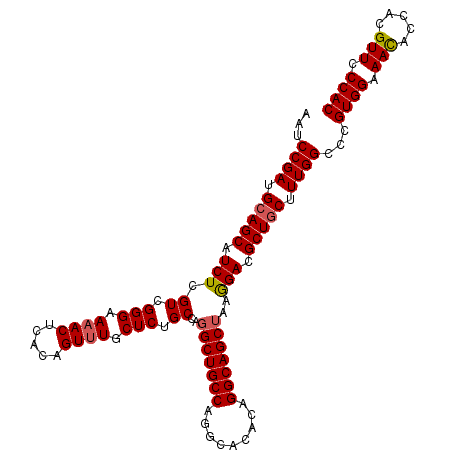
| Location | 4,806,579 – 4,806,689 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -42.34 |
| Consensus MFE | -40.50 |
| Energy contribution | -40.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4806579 110 - 20766785 GUGGGAACGUGGUGUUUCCACGGGCCAAAGCAGCGUCCUUAGCUGCCUGUGUGCCUGGCAGCCUGGCAGAGCAAACUGUGAGUUUUCCCGACGAGAUGCUGCAUCGGAUU ...((..(((((.....)))))..))...(((((((((...((((((.........))))))((.((((......)))).))..........).))))))))........ ( -44.00) >DroSec_CAF1 17877 110 - 1 GUGGGAACGUGGUGUUUCCACGGGCCAAAGCAGCGUCCUUAGCUGCCUGUGUGCCUGGCAGCCUGGCAGAGCAAACUGUGAGUUUUCCCGACGAGAUGCUGCAUCGGAUU ...((..(((((.....)))))..))...(((((((((...((((((.........))))))((.((((......)))).))..........).))))))))........ ( -44.00) >DroSim_CAF1 17847 110 - 1 GUGGGAACGUGGUGUUUCCACGGGCCAAAGCAGCGUCCUUAGCUGCCUGUGUGCCUGGCAGCCUGGCAGAGCAAACUGUGAGUUUUCCCGACGAGAUGCUGCAUCGGAUU ...((..(((((.....)))))..))...(((((((((...((((((.........))))))((.((((......)))).))..........).))))))))........ ( -44.00) >DroEre_CAF1 17816 110 - 1 GUGGGAACCUGGUAUUUCCACAGGCCAAAGCAGCGUCUUUUGCUGCCUGUGUGCUUGGCAGCCUGGCAGAGCAAACUGUGAGUUUUCCCGACGAGAUGCUCCAUCGGAUU (((((((.......))))))).((....((((.((((....((((((.........))))))((.((((......)))).)).......))))...))))))........ ( -35.10) >DroYak_CAF1 15542 110 - 1 GUGGGAACCUGGUGUUUCCACAGGCCAAAGCAGCGUCUUUAGCUGCCUGUGUGCUUGGCAGCCUGGCAGAGCAAACUGUGAGUGUUCCCGACGAGAUGCUGCAUCGGAUU ((((((((.....))))))))...((...((((((((((..((((((.........))))))((.((((......)))).))..........))))))))))...))... ( -44.60) >consensus GUGGGAACGUGGUGUUUCCACGGGCCAAAGCAGCGUCCUUAGCUGCCUGUGUGCCUGGCAGCCUGGCAGAGCAAACUGUGAGUUUUCCCGACGAGAUGCUGCAUCGGAUU ((((((((.....))))))))...((...((((((((....((((((.........))))))((.((((......)))).))............))))))))...))... (-40.50 = -40.90 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:03 2006