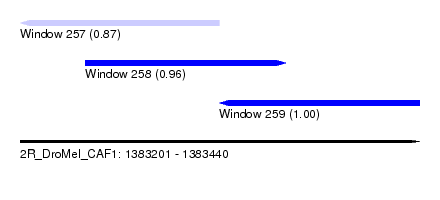
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,383,201 – 1,383,440 |
| Length | 239 |
| Max. P | 0.999562 |

| Location | 1,383,201 – 1,383,320 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -21.06 |
| Energy contribution | -20.98 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1383201 119 - 20766785 CAUCUUUGCCUUUUAUUUGGGCGCAUAAAAAUUUGCAAGCUAAUUUUUUUUAGCUAGGCAGUAAAAAGUAAAAAGUGGGUCCAAAUGAGAAACGAAGUCAGAAGGAGGACUGGUUGGCU- .......(((((((((((((((.(((......((((.((((((......))))))..)))).............))).))))))))))))(((..((((........)))).)))))).- ( -33.41) >DroSec_CAF1 10484 110 - 1 CAUCCUUGCCUUUUAUUUUGGCGCAUAAAAAUUUGCAAGCUAAUUU--UUUAGCUCGGCAGUAAAAAGUAAAAAGUGGGGCCAAAUGAGAAACGAAGUCAGAAGGA-------CUGGCU- ...(((..(.(((((((((.(((((........))).((((((...--.))))))..)).....))))))))).)..)))...............((((((.....-------))))))- ( -28.30) >DroSim_CAF1 15169 110 - 1 CAUCUUUGCCUUUUAUUUGGGCGCAUAAAAAUUUGCAAGCUAAUUU--UUUAGCUCGGCAGUAAAAAGUAAAAAGUGGGGCCAAAUGAGAAACGAAGUCAGAAGGA-------CUGGCU- .......(((((((((((((.(.(((......((((.((((((...--.))))))..)))).............))).).)))))))))).....((((.....))-------))))).- ( -29.81) >DroEre_CAF1 8294 111 - 1 CAUCUUUGGCUUUUAUUUGGGUGCAUAAAAAUUUGCAAGCUAAUUU--UUUAGCUCGGCUGUAAAAAAUAAAAAGUGGGACCAAAUGAAAAGCGAAGCAAGAGGGA-------UGAGAUU (((((((.((((((((((((.((((........))))((((((...--.))))))(.(((.............))).)..))))))).))))).)).......)))-------))..... ( -26.43) >DroYak_CAF1 6604 110 - 1 CAUCUUUGGCUUUUAUUUGGGUGCAUAAAGAUAUGCAAGCUAAUUU--UUUAGCUCGGCUGUAAAAAAUAAAAAGUAGGACCAAAUGAAAAACGAAGCCAGAAGGA-------CGGGGU- ....((((((((((((((((.((((((....))))))((((((...--.)))))).........................)))))))......)))))))))....-------......- ( -26.90) >consensus CAUCUUUGCCUUUUAUUUGGGCGCAUAAAAAUUUGCAAGCUAAUUU__UUUAGCUCGGCAGUAAAAAGUAAAAAGUGGGACCAAAUGAGAAACGAAGUCAGAAGGA_______CUGGCU_ ...(((((..((((((((((.((((........))).((((((......)))))).......................).))))))))))..)))))....................... (-21.06 = -20.98 + -0.08)

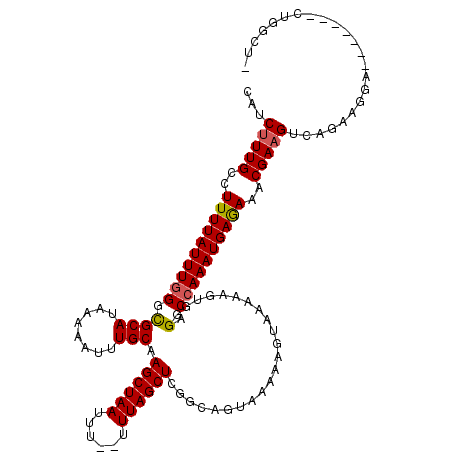
| Location | 1,383,240 – 1,383,360 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1383240 120 + 20766785 ACCCACUUUUUACUUUUUACUGCCUAGCUAAAAAAAAUUAGCUUGCAAAUUUUUAUGCGCCCAAAUAAAAGGCAAAGAUGUGAACAGAAAAAGUUUUGGAAUUGCUAUUCUUGGCAGAUC .((.(((((((.((.((((((((((((((((......)))))).(((........)))...........))))).....))))).)))))))))...))..((((((....))))))... ( -27.80) >DroSec_CAF1 10516 118 + 1 CCCCACUUUUUACUUUUUACUGCCGAGCUAAA--AAAUUAGCUUGCAAAUUUUUAUGCGCCAAAAUAAAAGGCAAGGAUGUGAACAGAAAAAGUUUUGGAAUUGCUAUUCUUGGCAGAUC ((..(((((((.((.(((((..(((((((((.--...)))))))(((........)))(((.........)))..))..))))).)))))))))...))..((((((....))))))... ( -29.80) >DroSim_CAF1 15201 117 + 1 CCCCACUUUUUACUUUUUACUGCCGAGCUAAA--AAAUUAGCUUGCAAAUUUUUAUGCGCCCAAAUAAAAGGCAAAGAUGUGAACGGA-AAAGUUUUGGAAUUGCUAUUCUUGGCAGAUC ((..(((((((....((((((((((((((((.--...)))))))(((........)))............)))).....)))))..))-)))))...))..((((((....))))))... ( -26.90) >DroEre_CAF1 8327 117 + 1 UCCCACUUUUUAUUUUUUACAGCCGAGCUAAA--AAAUUAGCUUGCAAAUUUUUAUGCACCCAAAUAAAAGCCAAAGAUGUGAACGGAA-AACUUUUGGAAUUGCCAUUCUUGGCAGAUC ..(((.(((((....((((((((.(((((((.--...)))))))))...(((((((........))))))).......))))))..)))-))....)))..((((((....))))))... ( -24.40) >DroYak_CAF1 6636 118 + 1 UCCUACUUUUUAUUUUUUACAGCCGAGCUAAA--AAAUUAGCUUGCAUAUCUUUAUGCACCCAAAUAAAAGCCAAAGAUGGGAACAGAAAAAGUUUUGGAUUUCCCAUUCUUGGCAGAAC .......((((((((.........(((((((.--...)))))))(((((....)))))....))))))))(((((..((((((((((((....)))))...)))))))..)))))..... ( -34.90) >consensus CCCCACUUUUUACUUUUUACUGCCGAGCUAAA__AAAUUAGCUUGCAAAUUUUUAUGCGCCCAAAUAAAAGGCAAAGAUGUGAACAGAAAAAGUUUUGGAAUUGCUAUUCUUGGCAGAUC .((.(((((((.((.(((((.((.(((((((......)))))))))...(((((((........)))))))........))))).)))))))))...))..((((((....))))))... (-21.70 = -22.38 + 0.68)

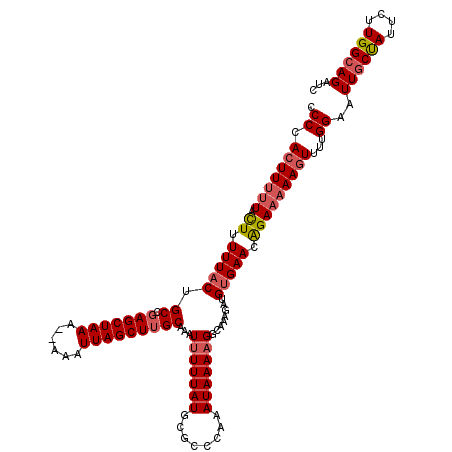
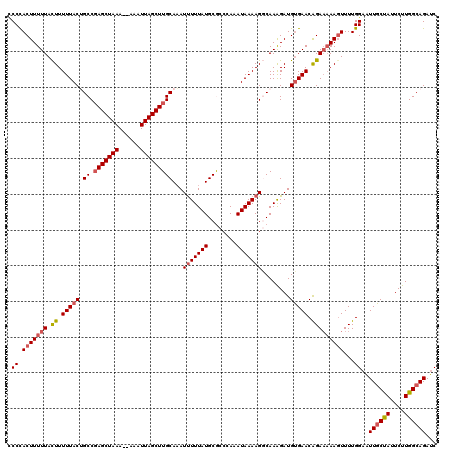
| Location | 1,383,320 – 1,383,440 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -27.01 |
| Energy contribution | -26.41 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1383320 120 - 20766785 CUUUGGCAUCAGCUCAAUUAUUCCAACUUGGUCUCGGCCAAGCAUUUGUUAAUAAUGGCUUAAAAUCAUAGGGAAUAGGAGAUCUGCCAAGAAUAGCAAUUCCAAAACUUUUUCUGUUCA ..((((((....(((...((((((..((((((....))))))((((.......))))..............)))))).)))...))))))((((((.((...........)).)))))). ( -30.70) >DroSec_CAF1 10594 120 - 1 CUUUGGCAUCAGCUCAAUUAUUCCAACUUGGUCUCGACCAAGCAUUUGUUAAUAAUGGUUUAAAAUCAUAGGAAAUAGGAGAUCUGCCAAGAAUAGCAAUUCCAAAACUUUUUCUGUUCA ..((((((....(((.....((((..((((((....))))))............(((((.....))))).))))....)))...))))))((((((.((...........)).)))))). ( -28.30) >DroSim_CAF1 15279 119 - 1 CUUUGGCAUCAGCUCAAUUAUUCCAACUUGGUCUCGGCCAAGCAUUUGUUAAUAAUGGCUUAAAAUCAUAGGAAAUAGGAGAUCUGCCAAGAAUAGCAAUUCCAAAACUUU-UCCGUUCA .(((((((....(((.....((((..((((((....))))))((((.......)))).............))))....)))...)))))))....................-........ ( -24.80) >DroEre_CAF1 8405 119 - 1 CUUUGGCAACAGCUCAAUUAUUCCAACUUGGCCUCGGCCAAGCAUUUCUUAAUAAUGGCUGAAAAUCAUAGGAAAUAGGGGAUCUGCCAAGAAUGGCAAUUCCAAAAGUU-UUCCGUUCA ....(((..(((((..((((......((((((....)))))).......))))...))))).........(((((....(((..(((((....)))))..)))......)-))))))).. ( -33.72) >DroYak_CAF1 6714 120 - 1 CCUUGGCAACAGCUCAAUUAUUCCAACAUGGCCUCGGCCAGGCAUUUUUUAAUAAUGGCUAAAAAUCAUCGGAAAUAGGAGUUCUGCCAAGAAUGGGAAAUCCAAAACUUUUUCUGUUCC ..((((((..(((((.....((((.....(((....)))((.((((.......)))).))..........))))....))))).))))))((((((((((.........)))))))))). ( -33.80) >consensus CUUUGGCAUCAGCUCAAUUAUUCCAACUUGGUCUCGGCCAAGCAUUUGUUAAUAAUGGCUUAAAAUCAUAGGAAAUAGGAGAUCUGCCAAGAAUAGCAAUUCCAAAACUUUUUCUGUUCA ..((((((....(((.....((((..((((((....))))))((((.......)))).............))))....)))...))))))((((((.................)))))). (-27.01 = -26.41 + -0.60)

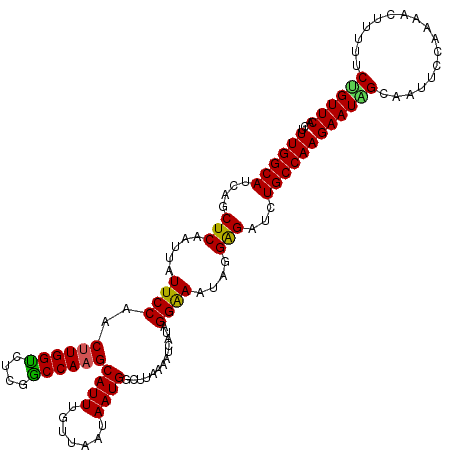
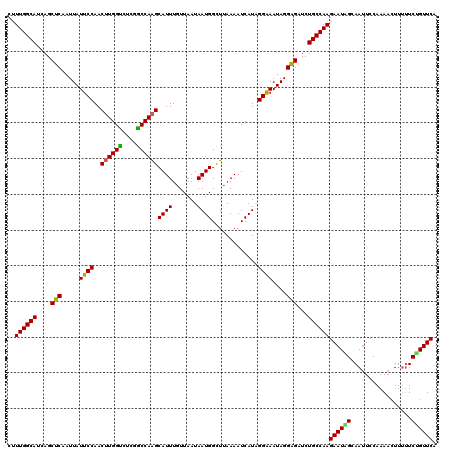
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:57 2006