
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,805,251 – 4,805,444 |
| Length | 193 |
| Max. P | 0.996020 |

| Location | 4,805,251 – 4,805,371 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -35.32 |
| Energy contribution | -35.84 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4805251 120 + 20766785 CUAAACUACAGGCACUCUGCUCCAUCUAUGUGAAUUUUUCAUAUGACAACCUUUGGCGAGUGUAUCUAGCAAGGCGUAUCGCUGCGGAUGCUCCGCACUCGCGUCCGCAUCCGCUAGAGA ...........((((((.(((.....(((((((.....))))))).........))))))))).((((((..((((...))))((((((((.........))))))))....)))))).. ( -39.84) >DroSec_CAF1 16554 120 + 1 CUAAGCUACAGGCACUCUGCUCCAUCUAUGUGAAUUUUUCAUAUGACAACCUUUGGCGAGUGUAUCUAGCAAGGCGUAUCGCUGCGGAUGCUCCGCACUCGCGUCCGCAUCCGCUAGAGG ....((((...((((((.(((.....(((((((.....))))))).........)))))))))...))))..((((...))))((((((((..(((....)))...))))))))...... ( -40.64) >DroSim_CAF1 16520 120 + 1 CUAAGCUACAGGCACUCUGCUCCAUCUAUGUGAAUUUUUCAUAUGACAACCUUUGGCGAGUGUAUCUAGCAAGGCGUAUCGCUGCGGAUGCUCCGCACUCGCGUCCGCAUCCGCUAGAGG ....((((...((((((.(((.....(((((((.....))))))).........)))))))))...))))..((((...))))((((((((..(((....)))...))))))))...... ( -40.64) >DroEre_CAF1 16487 120 + 1 CUAAAGUAUAGGCACACUGCUCCAUCUAUCUGAGUUUUUCAUAUGACAACCUUUGGCGAGUGUAUCUAGCAAGGCGUAUCGCUGCGGAUGCUCCGCACUCGCGUCCGCAUCCGCUAGAGG .............(((((.(.(((((.....))(((........)))......))).)))))).((((((..((((...))))((((((((.........))))))))....)))))).. ( -35.20) >DroYak_CAF1 14243 117 + 1 CUAAACUACA--CACUCUGCUCCAUCUUUUUGAAUUUUUCAUAUGAUAACCUUUGGCGAGUGUAUCUAGCAAGGCGUAUCGUUGCGGAUGUUUCGCACUCGCGUCCGCAUCCGCUAGAG- .........(--(((((.(((..(((....(((.....)))...))).......))))))))).((((((...(((......)))((((((..(((....)))...)))))))))))).- ( -34.30) >consensus CUAAACUACAGGCACUCUGCUCCAUCUAUGUGAAUUUUUCAUAUGACAACCUUUGGCGAGUGUAUCUAGCAAGGCGUAUCGCUGCGGAUGCUCCGCACUCGCGUCCGCAUCCGCUAGAGG ...........((((((.(((.....((((((((...)))))))).........))))))))).((((((..((((...))))((((((((.........))))))))....)))))).. (-35.32 = -35.84 + 0.52)

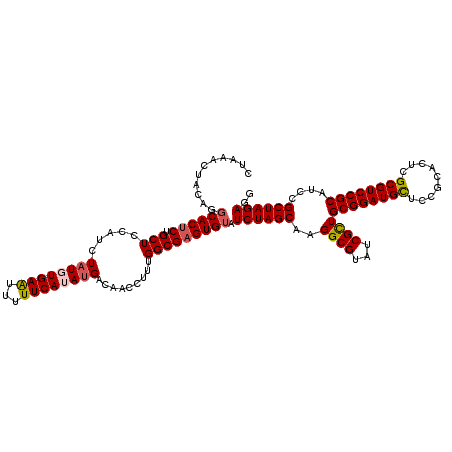
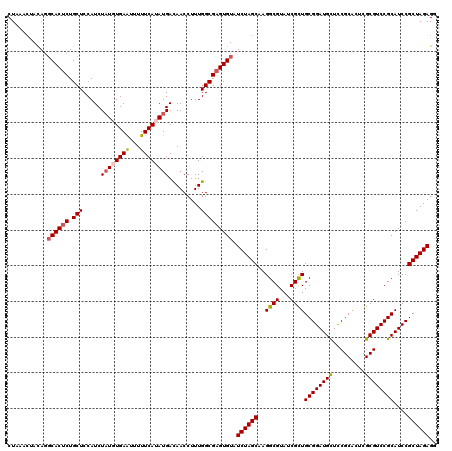
| Location | 4,805,291 – 4,805,407 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 98.02 |
| Mean single sequence MFE | -39.46 |
| Consensus MFE | -38.08 |
| Energy contribution | -38.12 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4805291 116 - 20766785 CAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUUCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGCGAUACGCCUUGCUAGAUACACUCGCCAAAGGUUGUCAUAU ...(((((((.((..(((((...))))).)).)))))))...((((((((...(((....)))..))))))))...((((.(((((((.((.....)).))..))))))))).... ( -41.70) >DroSec_CAF1 16594 116 - 1 CAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUCCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGCGAUACGCCUUGCUAGAUACACUCGCCAAAGGUUGUCAUAU ...(((((((.((..(((((...))))).)).)))..)))).((((((((...(((....)))..))))))))...((((.(((((((.((.....)).))..))))))))).... ( -39.20) >DroSim_CAF1 16560 116 - 1 CAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUCCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGCGAUACGCCUUGCUAGAUACACUCGCCAAAGGUUGUCAUAU ...(((((((.((..(((((...))))).)).)))..)))).((((((((...(((....)))..))))))))...((((.(((((((.((.....)).))..))))))))).... ( -39.20) >DroEre_CAF1 16527 116 - 1 CAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUCCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGCGAUACGCCUUGCUAGAUACACUCGCCAAAGGUUGUCAUAU ...(((((((.((..(((((...))))).)).)))..)))).((((((((...(((....)))..))))))))...((((.(((((((.((.....)).))..))))))))).... ( -39.20) >DroYak_CAF1 14281 115 - 1 CAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUU-CUCUAGCGGAUGCGGACGCGAGUGCGAAACAUCCGCAACGAUACGCCUUGCUAGAUACACUCGCCAAAGGUUAUCAUAU ...(((((((.((..(((((...))))).)).)).)-)))).(((((((((.((....)).))...)))))))...((((.(((((((.((.....)).))..))))))))).... ( -38.00) >consensus CAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUCCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGCGAUACGCCUUGCUAGAUACACUCGCCAAAGGUUGUCAUAU ...(((((((.((..(((((...))))).)).)))..)))).((((((((...(((....)))..))))))))...((((.(((((((.((.....)).))..))))))))).... (-38.08 = -38.12 + 0.04)

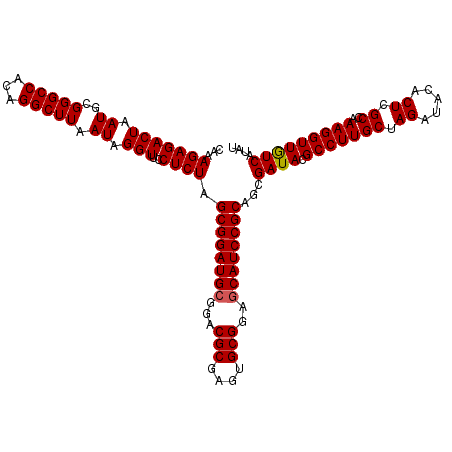
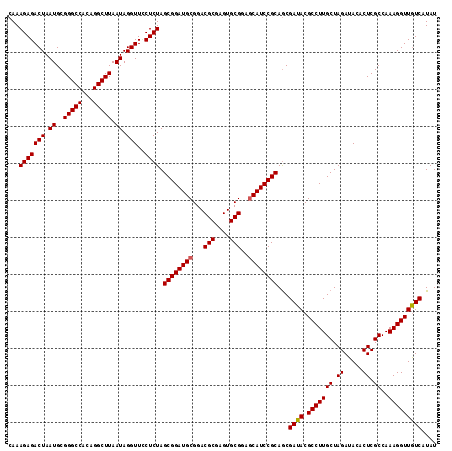
| Location | 4,805,331 – 4,805,444 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.20 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -38.66 |
| Energy contribution | -38.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4805331 113 - 20766785 --ACACAUAUGCAGAGCAGGAGCUUGAGGGACACGCACACAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUUCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGC --........((.((((....))))(((..((.((((..............))))((((...))))......))..)))..((((((((...(((....)))..)))))))).)) ( -39.24) >DroSec_CAF1 16634 114 - 1 -CACACAUAUGCAGAGCAGGAGCUUGAGGGACACGCACACAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUCCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGC -.........((.((((....))))(((((((.((((..............))))((((...))))......)))))))..((((((((...(((....)))..)))))))).)) ( -41.84) >DroSim_CAF1 16600 115 - 1 ACACACACAUGCAGAGCAGGAGCUUGAGGGACACGCACACAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUCCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGC ..........((.((((....))))(((((((.((((..............))))((((...))))......)))))))..((((((((...(((....)))..)))))))).)) ( -41.84) >DroEre_CAF1 16567 115 - 1 AUACACACAUGCAGAGCAGGAGCUUGAGGGACACGCACACAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUCCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGC ..........((.((((....))))(((((((.((((..............))))((((...))))......)))))))..((((((((...(((....)))..)))))))).)) ( -41.84) >DroYak_CAF1 14321 109 - 1 -----CACUUGCAGAGCAGGAGCUUGAGGGACACGCACACAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUU-CUCUAGCGGAUGCGGACGCGAGUGCGAAACAUCCGCAAC -----.......(((((..(((((((..((...((((..............))))..)).))))))).....)))-))...(((((((((.((....)).))...)))))))... ( -38.24) >consensus __ACACACAUGCAGAGCAGGAGCUUGAGGGACACGCACACAAAGAGACUAAUGCGGGCCACAGGCUUAAUAGGUUCCUCUAGCGGAUGCGGACGCGAGUGCGGAGCAUCCGCAGC .............((((....))))(((((((.((((..............))))((((...))))......)))))))..((((((((...(((....)))..))))))))... (-38.66 = -38.94 + 0.28)

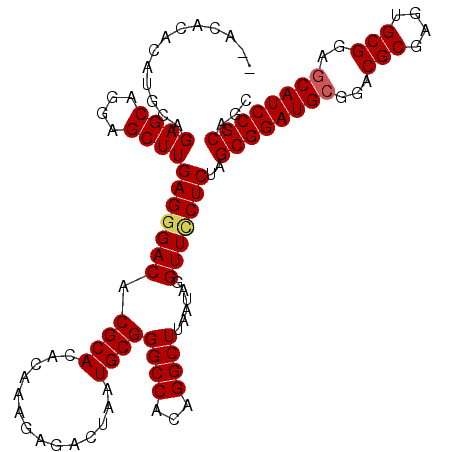
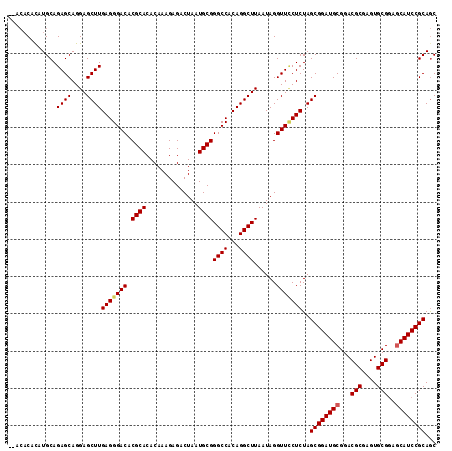
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:58 2006