| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,719,877 – 4,719,985 |
| Length | 108 |
| Max. P | 0.909508 |

| Location | 4,719,877 – 4,719,985 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
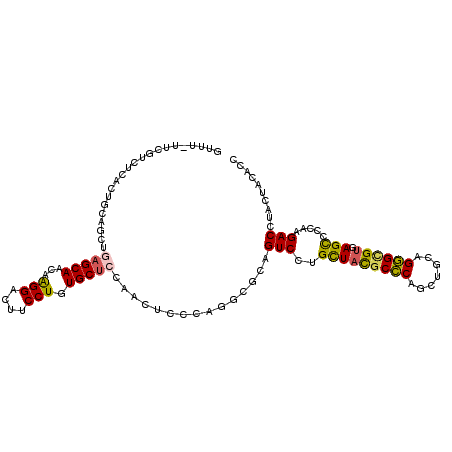
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -20.69 |
| Energy contribution | -20.25 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4719877 108 + 20766785 GUUU--GUUCCCCAACUCAGCUGAGCAGCAAGGACUUCCUGUGCUCCAACUCCCAGGCGCAGUCCUGCUACGCCCAGCUGCAGGGAGUGAGCCCCAAGACCUACUACACC (((.--.(((((.....((((((.((((((.(((((....(((((..........))))))))))))))..)).))))))..)))))..))).................. ( -38.80) >DroVir_CAF1 298 109 + 1 GUCUGUCUAUU-UUAUCUAGCUGAGCAAUAGGGAUUUCCUGUGCUCCAAUGGCGAGGCGCAGUCAUGUUACGCUCAGCUACAGAGCGUUAGCCCCAAGACGUACUACACC ((.(((((...-.......((((((((...(((....))).)))))....)))(.((((((....))).((((((.......))))))..))).).))))).))...... ( -32.60) >DroPse_CAF1 1830 110 + 1 GCUUUUUCGUCUGACUGCAGCUGAGCACCAAGGACUUCCUAUGCUCAAAUUCCCAGUCGCAGUCUUGCUACGCCCAGCUGCAGGGCGUGAGUCCCAAGACGUACUACACC .......(((((...((((((((((((...(((....))).))))).........((.((......)).))....)))))))((((....))))..)))))......... ( -37.40) >DroMoj_CAF1 310 110 + 1 GUCUGUCUCUCUCUCUCCAGUUAAGCAAUAAGGAUUUCCUGUGCUCCACCAACGAGGCCCAGUCCUGCUUUGCUCAGCUGCAGAGCGUCAGCCCCAAGACCUAUUACACA ((((..((..(.((((.(((((.(((((..((((....(((.((..(......)..)).)))))))...))))).))))).)))).)..)).....)))).......... ( -31.40) >DroAna_CAF1 296 106 + 1 AU----UCUUCUUAUUGCAGUUGAGCAGCAAGGACUUCCUGUGCAGCAACUCCGAGUCGCAGUCGUGCUAUGCCCAGUUACAGGGUGUUAGCCCCAAGACCUACUACACC ..----........(((.(((((....((((((....))).)))..))))).)))......(((..(((((((((.......))))).)))).....))).......... ( -23.90) >DroPer_CAF1 3409 110 + 1 GCUUUUUCGUCUGACUGCAGCUGAGCACCAAGGACUUCCUGUGCUCAAAUUCCCAGUCGCAGUCCUGCUACGCCCAGCUGCAGGGCGUGAGUCCCAAGACGUAUUACACC .......(((((...(((((((((((((..(((....))))))))).........((.(((....))).))....)))))))((((....))))..)))))......... ( -41.70) >consensus GUUU_UUCGUCUCACUGCAGCUGAGCAACAAGGACUUCCUGUGCUCCAACUCCCAGGCGCAGUCCUGCUACGCCCAGCUGCAGGGCGUGAGCCCCAAGACCUACUACACC ......................(((((...(((....))).)))))...............(((..(((((((((.......)))))).))).....))).......... (-20.69 = -20.25 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:41 2006