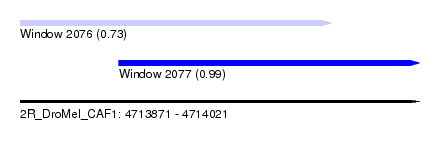
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,713,871 – 4,714,021 |
| Length | 150 |
| Max. P | 0.989954 |

| Location | 4,713,871 – 4,713,988 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -17.00 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4713871 117 + 20766785 AUUUUAAAAUCAGUUCUUCAAUU---AAGCCGUAUCUUAGAGAUUCAUUAUCUUGUGUCUUAGAGAUUUAUCAGCUGUAUCUUUAAUCAGAUUAAUCAGUUGUAUCUUAGAGAUUCAUCG .......................---.............(((((.....)))))(.(((((.(((((....((((((.((((......))))....)))))).))))).))))).).... ( -20.10) >DroSec_CAF1 7380 104 + 1 ------------GUUCU-CAUUUAAGAAGCUGUAUCUUAGAGAUUCAUUAUCUUGUAUCUUAGAGAUUCACCAGCGUUAUCCUAG---AGAUUCUUUAGCUGUAUCUUCGAGAUUUAUCA ------------..(((-(....((((.((((.(((((.(((((.((......)).))))).)))))....))))......((((---((...)))))).....)))).))))....... ( -22.00) >DroSim_CAF1 6693 104 + 1 ------------GUUCU-UAUUUCAGAAGCUGUAUCUCAGAGAUUCAUUAUCUUGUAUCUUAAAGAUUCACCAGCUGUAUCCUAG---AGAUUCUUUAGCUGUAUUUUCGAGAUUCAUCG ------------.((((-......))))(.((.(((((.(((((.((......)).)))))...((.....((((((.(((....---.)))....)))))).....))))))).)).). ( -20.00) >consensus ____________GUUCU_CAUUU_AGAAGCUGUAUCUUAGAGAUUCAUUAUCUUGUAUCUUAGAGAUUCACCAGCUGUAUCCUAG___AGAUUCUUUAGCUGUAUCUUCGAGAUUCAUCG ............................(.((.(((((.(((((.((......)).))))).(((((....((((((.(((........)))....)))))).))))).))))).)).). (-17.00 = -16.57 + -0.43)

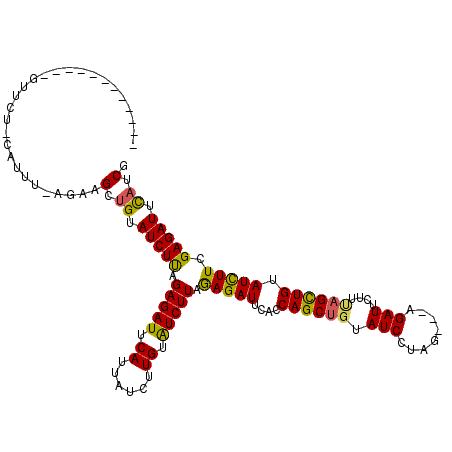
| Location | 4,713,908 – 4,714,021 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.03 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4713908 113 + 20766785 AGAUUCAUUAUCUUGUGUCUUAGAGAUUUAUCAGCUGUAUCUUUAAUCAGAUUAAUCAGUUGUAUCUUAGAGAUUCAUCGGAAUAGUGCAGUCAAUUCAUGAUUAAAUUUGCA .((((((((((((((.(((((.(((((....((((((.((((......))))....)))))).))))).))))).))..)).)))))).)))).................... ( -26.20) >DroSec_CAF1 7407 110 + 1 AGAUUCAUUAUCUUGUAUCUUAGAGAUUCACCAGCGUUAUCCUAG---AGAUUCUUUAGCUGUAUCUUCGAGAUUUAUCAGAAUAGUGCAGUCAAAUUAUUAUUAAAGAUUUA .((((((((((..((.((((..(((((....((((...(((....---.)))......)))).)))))..))))....))..)))))).)))).................... ( -23.40) >DroSim_CAF1 6720 110 + 1 AGAUUCAUUAUCUUGUAUCUUAAAGAUUCACCAGCUGUAUCCUAG---AGAUUCUUUAGCUGUAUUUUCGAGAUUCAUCGGAAUAAUGCAGACAAUUUAUGAUUAAAGAUUCA .((.((...(((((........)))))(((..((.(((...((((---((...))))))((((((((((((......)))))..)))))))))).))..))).....)).)). ( -19.30) >consensus AGAUUCAUUAUCUUGUAUCUUAGAGAUUCACCAGCUGUAUCCUAG___AGAUUCUUUAGCUGUAUCUUCGAGAUUCAUCGGAAUAGUGCAGUCAAUUUAUGAUUAAAGAUUCA .((((((((((..((.((((..(((((....((((((.(((........)))....)))))).)))))..))))....))..)))))).)))).................... (-22.46 = -22.03 + -0.43)

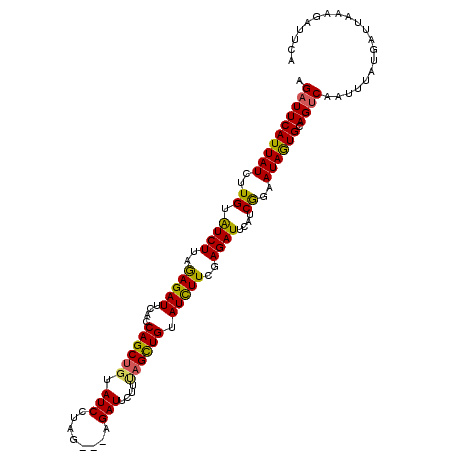
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:37 2006