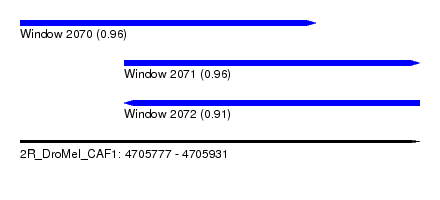
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,705,777 – 4,705,931 |
| Length | 154 |
| Max. P | 0.956562 |

| Location | 4,705,777 – 4,705,891 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.92 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4705777 114 + 20766785 GCUCGUAAAAGAUUUCAAACACCCAAACGUGACCAAUGCCAAUUUGCGCUCCAAUCAAUGGACGCCUCGUCACUUAUGGCCAAAAAUUUGGUCAAUUACGAUCAAUU------AAGCCAU (.((((((....................(((((....((......))((((((.....)))).))...)))))...(((((((....))))))).)))))).)....------....... ( -26.20) >DroSec_CAF1 15492 114 + 1 GCUCGUAAAAGAUUUCAAACAUCCAAACGUGACCAAUGCCAAUUUGCGCUCCAAUCAAUGGACACCUCGACACUUAUGGCCAAAAAUUUGGUCAACUACGAUCAAUU------AAGCCAC (.(((((.....................(((......((......))(.((((.....))))).......)))...(((((((....)))))))..))))).)....------....... ( -18.60) >DroSim_CAF1 15821 114 + 1 GCUCGUAAAAGAUUUCAAACACCCAAACGUGACCAAUGCCAAUUUGCGCUCCAAUCAAUGGACGCCUCGACACUUAUGGCCAAAAAUUUGGUCAAUUACGAUCAAUU------AAGCCAU (.(((((((((........(((......)))..............(((.((((.....))))))).......))).(((((((....))))))).)))))).)....------....... ( -22.90) >DroEre_CAF1 15216 114 + 1 GCUCGUAAAAGAUUUCAAACACCGAAACGUGACCAAUGCCAAUUUUCGCUCCAAUCAAUGGACGCCUCGUCACUUAUGGCCAAAAAUUUGGUCAAUUACGAUCAAUU------AAGCCAU (.((((((....................(((((.(((....)))...((((((.....)))).))...)))))...(((((((....))))))).)))))).)....------....... ( -23.40) >DroYak_CAF1 15321 120 + 1 GCUCGUAAAAGAUUUCAAACACCCAAACGUGACCAAUGCCAAUUCGCGCUCCAAUCAAUGGACGCCUCUUCACUUAUGGCCAAAAAUUUGGUCAAUUAUGAUCAAUUACGAUCAAGCCAU (.((((((..((((..............((((.....((......))((((((.....)))).))....))))...(((((((....))))))).....))))..)))))).)....... ( -25.20) >consensus GCUCGUAAAAGAUUUCAAACACCCAAACGUGACCAAUGCCAAUUUGCGCUCCAAUCAAUGGACGCCUCGUCACUUAUGGCCAAAAAUUUGGUCAAUUACGAUCAAUU______AAGCCAU (.((((((....................(((((....((......))((((((.....)))).))...)))))...(((((((....))))))).)))))).)................. (-19.88 = -20.92 + 1.04)

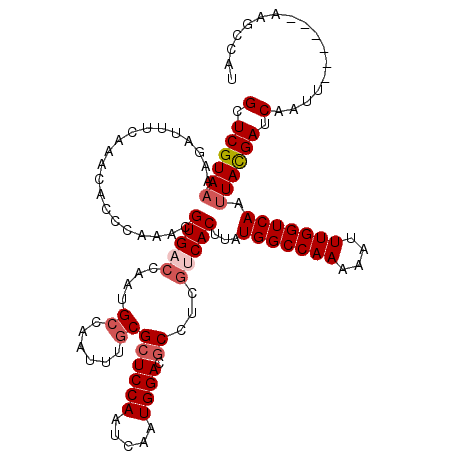
| Location | 4,705,817 – 4,705,931 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -21.59 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4705817 114 + 20766785 AAUUUGCGCUCCAAUCAAUGGACGCCUCGUCACUUAUGGCCAAAAAUUUGGUCAAUUACGAUCAAUU------AAGCCAUACACAUGCAAAUUAUUGCCACCUGCUGCGCGAAGAACGAU ..(((((((..((....(((((((...))))...((((((.......((((((......))))))..------..))))))..)))((((....))))....))..)))))))....... ( -28.50) >DroSec_CAF1 15532 114 + 1 AAUUUGCGCUCCAAUCAAUGGACACCUCGACACUUAUGGCCAAAAAUUUGGUCAACUACGAUCAAUU------AAGCCACACACAUGCAAAUUAUUGCCACCUGCUGGGCGAAGAACGAU ((((((((.((((.....))))).............((((.......((((((......))))))..------..)))).......))))))).(((((........)))))........ ( -24.70) >DroSim_CAF1 15861 114 + 1 AAUUUGCGCUCCAAUCAAUGGACGCCUCGACACUUAUGGCCAAAAAUUUGGUCAAUUACGAUCAAUU------AAGCCAUACACAUGCAAAUUAUUGCCACCUGCUGGGCAAAGAACGAU (((((((((((((.....)))).)).........((((((.......((((((......))))))..------..)))))).....))))))).(((((........)))))........ ( -29.70) >DroEre_CAF1 15256 114 + 1 AAUUUUCGCUCCAAUCAAUGGACGCCUCGUCACUUAUGGCCAAAAAUUUGGUCAAUUACGAUCAAUU------AAGCCAUACACAUGCAAAUUAUUGCCACCUGCUGCGUGAAAAGCAAU ..(((((((((((.....)))).((.........((((((.......((((((......))))))..------..)))))).....((((....))))........)))))))))..... ( -24.30) >DroYak_CAF1 15361 120 + 1 AAUUCGCGCUCCAAUCAAUGGACGCCUCUUCACUUAUGGCCAAAAAUUUGGUCAAUUAUGAUCAAUUACGAUCAAGCCAUACACAUGCAAAUGAUUGCCACCUGCUGCGUGAAAAACGAU ..(((((((..((....((((.(.............(((((((....)))))))....(((((......))))).)))))......((((....))))....))..)))))))....... ( -29.40) >consensus AAUUUGCGCUCCAAUCAAUGGACGCCUCGUCACUUAUGGCCAAAAAUUUGGUCAAUUACGAUCAAUU______AAGCCAUACACAUGCAAAUUAUUGCCACCUGCUGCGCGAAGAACGAU ..(((((((((((.....)))).((.........((((((.......((((((......))))))..........)))))).....((((....)))).....)).)))))))....... (-21.59 = -22.15 + 0.56)

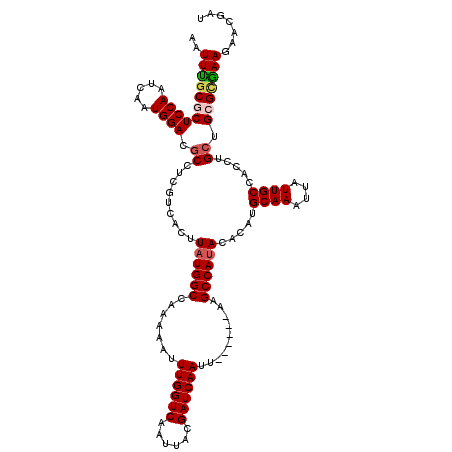

| Location | 4,705,817 – 4,705,931 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -28.46 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4705817 114 - 20766785 AUCGUUCUUCGCGCAGCAGGUGGCAAUAAUUUGCAUGUGUAUGGCUU------AAUUGAUCGUAAUUGACCAAAUUUUUGGCCAUAAGUGACGAGGCGUCCAUUGAUUGGAGCGCAAAUU .((((((((.(((((((((((.......)))))).)))))((((((.------((((..(((....)))...))))...))))))))).))))).(((((((.....)))).)))..... ( -34.10) >DroSec_CAF1 15532 114 - 1 AUCGUUCUUCGCCCAGCAGGUGGCAAUAAUUUGCAUGUGUGUGGCUU------AAUUGAUCGUAGUUGACCAAAUUUUUGGCCAUAAGUGUCGAGGUGUCCAUUGAUUGGAGCGCAAAUU .........(((((((..(((((((....(((((((...(((((((.------((((..(((....)))...))))...))))))).))).)))).)).)))))..)))).)))...... ( -31.50) >DroSim_CAF1 15861 114 - 1 AUCGUUCUUUGCCCAGCAGGUGGCAAUAAUUUGCAUGUGUAUGGCUU------AAUUGAUCGUAAUUGACCAAAUUUUUGGCCAUAAGUGUCGAGGCGUCCAUUGAUUGGAGCGCAAAUU .(((....((((((.....).)))))......((((...(((((((.------((((..(((....)))...))))...))))))).))))))).(((((((.....)))).)))..... ( -30.10) >DroEre_CAF1 15256 114 - 1 AUUGCUUUUCACGCAGCAGGUGGCAAUAAUUUGCAUGUGUAUGGCUU------AAUUGAUCGUAAUUGACCAAAUUUUUGGCCAUAAGUGACGAGGCGUCCAUUGAUUGGAGCGAAAAUU ....(((.(((((((((((((.......)))))).))).(((((((.------((((..(((....)))...))))...))))))).)))).)))((.((((.....))))))....... ( -31.50) >DroYak_CAF1 15361 120 - 1 AUCGUUUUUCACGCAGCAGGUGGCAAUCAUUUGCAUGUGUAUGGCUUGAUCGUAAUUGAUCAUAAUUGACCAAAUUUUUGGCCAUAAGUGAAGAGGCGUCCAUUGAUUGGAGCGCGAAUU .((.((((((((((((((((((.....))))))).))).((((((.((((((....))))))........(((....))))))))).))))))))(((((((.....)))).)))))... ( -39.40) >consensus AUCGUUCUUCGCGCAGCAGGUGGCAAUAAUUUGCAUGUGUAUGGCUU______AAUUGAUCGUAAUUGACCAAAUUUUUGGCCAUAAGUGACGAGGCGUCCAUUGAUUGGAGCGCAAAUU ....(((.((((...(((.(((.(((....)))))).)))((((((.........(((.(((....))).)))......))))))..)))).)))(((((((.....)))).)))..... (-28.46 = -27.78 + -0.68)

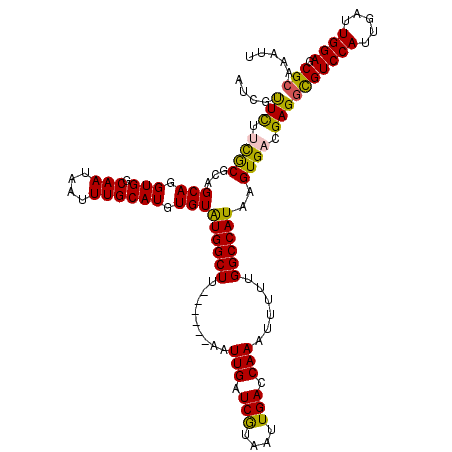
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:32 2006