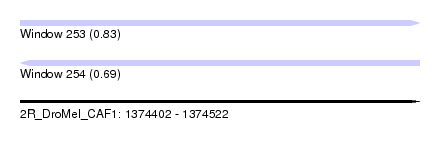
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,374,402 – 1,374,522 |
| Length | 120 |
| Max. P | 0.825096 |

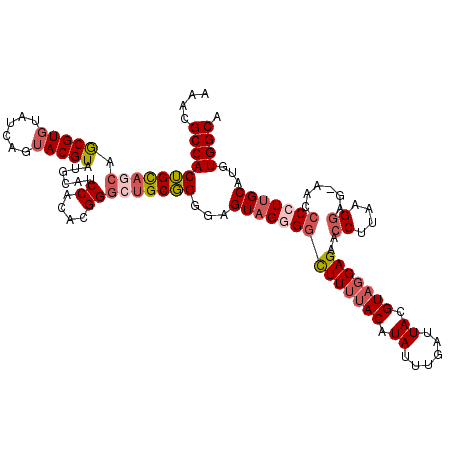
| Location | 1,374,402 – 1,374,522 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -39.54 |
| Consensus MFE | -28.52 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.825096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1374402 120 + 20766785 UGCCACAUGCAAGGGGUUCCUCCUUAAGGUUCUCCUACGUAAUCAAAUAUGUAAAAGCCCCUACUUCCCGCAGCCCGUGUGGAUGCAUACGUACUGAUACACGCUGCUGCAGUGGCGUUU .(((((.((((((((((((((.....)))......((((((......))))))..))))))).......(((((..(((((....)))))(((....)))..)))))))))))))).... ( -42.40) >DroSec_CAF1 6678 119 + 1 UGCCACAUGCAAGGGGUUCCUCCUUAAGGUUCUCCUACGUAAUCAAAUAUGUAAAAGCC-CUACUCCCCGCCGCCCGUGUGGAUGCAUACGUACUGAUACACGCUGCUGCAGUGGCGUUU .(((((..((..((((..(((.....)))..))))((((((......))))))...)).-.........((.((.((((((..(((....)))....))))))..)).)).))))).... ( -35.40) >DroSim_CAF1 11364 118 + 1 UGCCACAUGCAAGGGGUU-CUCCUUAAGGUUCUCCUACGUAAUCAAAUAUGUAAAAGCC-CUACUCCCCGCAGCCCGUGUGGAUGCAUACGUACUGAUACACGCUGCUGCAGUGGCGUUU .(((((.((((.((((..-.......(((..((..((((((......))))))..)).)-))...))))(((((..(((((....)))))(((....)))..)))))))))))))).... ( -39.60) >DroEre_CAF1 4703 117 + 1 UGCCACAUGCAAGGGGUU---CCUUAAGGUUCUCCUACGUAAUCAAAUAUGUAAAAGCCCCUACUCCCCGCAGCCCUUGUGGAUGCAUACGUACUGAUACACGUUGGUGCAGUGGCGUUU .(((((((((((((((((---......((....))((((((......))))))..))))))).....((((((...)))))).)))))..((((..((....))..)))).))))).... ( -42.00) >DroYak_CAF1 3137 117 + 1 AGCCACAUGCAAGGGGUU---GCUUAAGGUUCUCAUACGUAAUCAAAUAUGUGAAAAACCCAACCUCCAGCAGACCGUGUGGAUGCAUACGUACUGAUAAACGUUGGUACCGUGGCGUUU .(((((.(((..((((((---(.....((((.(((((..(......)..)))))..)))))))))))..)))....(((((....)))))((((..((....))..)))).))))).... ( -38.30) >consensus UGCCACAUGCAAGGGGUU_CUCCUUAAGGUUCUCCUACGUAAUCAAAUAUGUAAAAGCCCCUACUCCCCGCAGCCCGUGUGGAUGCAUACGUACUGAUACACGCUGCUGCAGUGGCGUUU .(((((.((((.((((...........((....))((((((......))))))............)))).(((..((((((....))))))..)))...........))))))))).... (-28.52 = -29.32 + 0.80)

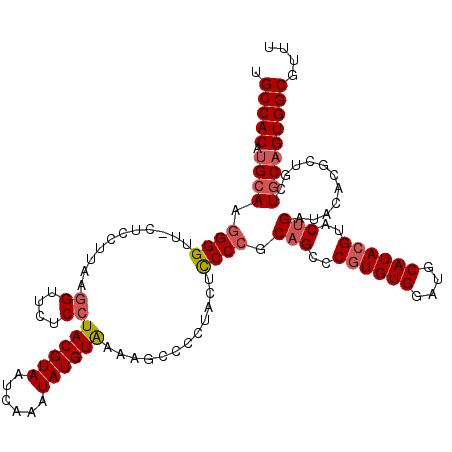
| Location | 1,374,402 – 1,374,522 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -29.30 |
| Energy contribution | -30.82 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1374402 120 - 20766785 AAACGCCACUGCAGCAGCGUGUAUCAGUACGUAUGCAUCCACACGGGCUGCGGGAAGUAGGGGCUUUUACAUAUUUGAUUACGUAGGAGAACCUUAAGGAGGAACCCCUUGCAUGUGGCA ....(((((((((((..(((((....(((....)))....))))).)))))))...((((((((((((((.((......)).)))))))..((((...))))...)))))))...)))). ( -43.90) >DroSec_CAF1 6678 119 - 1 AAACGCCACUGCAGCAGCGUGUAUCAGUACGUAUGCAUCCACACGGGCGGCGGGGAGUAG-GGCUUUUACAUAUUUGAUUACGUAGGAGAACCUUAAGGAGGAACCCCUUGCAUGUGGCA ....(((((((((((.((((((((......))))))..((....)))).))((((.....-..(((((((.((......)).)))))))..((((...))))..)))).)))).))))). ( -39.50) >DroSim_CAF1 11364 118 - 1 AAACGCCACUGCAGCAGCGUGUAUCAGUACGUAUGCAUCCACACGGGCUGCGGGGAGUAG-GGCUUUUACAUAUUUGAUUACGUAGGAGAACCUUAAGGAG-AACCCCUUGCAUGUGGCA ....((((((((((((((((((((......))))))..((....)))))))((((..(((-(((((((((.((......)).)))))))..))))).....-..)))).)))).))))). ( -43.70) >DroEre_CAF1 4703 117 - 1 AAACGCCACUGCACCAACGUGUAUCAGUACGUAUGCAUCCACAAGGGCUGCGGGGAGUAGGGGCUUUUACAUAUUUGAUUACGUAGGAGAACCUUAAGG---AACCCCUUGCAUGUGGCA ....(((((((((...(((((......)))))..((((((....))).)))((((...(((..(((((((.((......)).)))))))..))).....---..)))).)))).))))). ( -41.00) >DroYak_CAF1 3137 117 - 1 AAACGCCACGGUACCAACGUUUAUCAGUACGUAUGCAUCCACACGGUCUGCUGGAGGUUGGGUUUUUCACAUAUUUGAUUACGUAUGAGAACCUUAAGC---AACCCCUUGCAUGUGGCU ....((((((((.(((((.((((.(((..(((.((....)).)))..))).)))).))))).....((.(((((........))))).)))))....((---((....))))..))))). ( -35.30) >consensus AAACGCCACUGCAGCAGCGUGUAUCAGUACGUAUGCAUCCACACGGGCUGCGGGGAGUAGGGGCUUUUACAUAUUUGAUUACGUAGGAGAACCUUAAGGAG_AACCCCUUGCAUGUGGCA ....(((((((((((.(((((......)))))......((....)))))))))...((((((((((((((.((......)).)))))))..((....))......)))))))...)))). (-29.30 = -30.82 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:52 2006