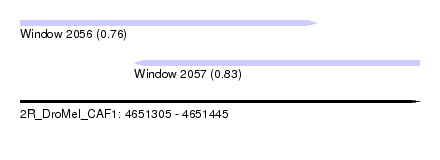
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,651,305 – 4,651,445 |
| Length | 140 |
| Max. P | 0.830156 |

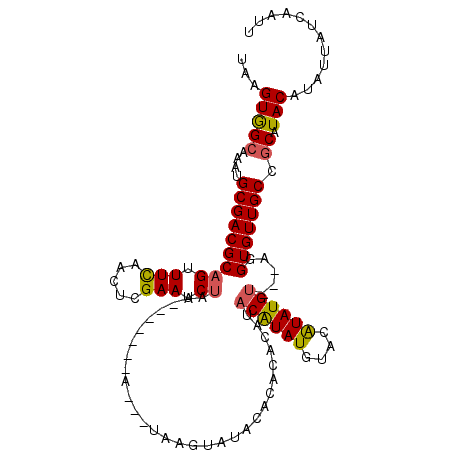
| Location | 4,651,305 – 4,651,409 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -15.44 |
| Energy contribution | -18.12 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4651305 104 + 20766785 AUCAGCGAUCUGCUUAGCCAGUGAACUGUUCUGAACUGCUAAUUGAUAAUAUGUAUGCGGCAACACCU---ACAUAUGUACAUACGUAUGUGUAUACUUACAUA---U-------UU .....((((..((((((.(((....)))..))))...))..)))).......(((((((....)....---((((((((....)))))))))))))).......---.-------.. ( -23.00) >DroSec_CAF1 5703 104 + 1 AUCAGCGAUCUGCUUAGCCAGUGAACUGUUCUGAACUGCUAAUUGAUAAUAUGUAUGCGGCAACACCU---ACAUACAUACAUAUGUAUGUAUGUGUAUACUUA---U-------UU .....((((..((((((.(((....)))..))))...))..))))..((((.(((((((....)....---((((((((((....)))))))))))))))).))---)-------). ( -29.00) >DroSim_CAF1 5988 104 + 1 AUCAGCGAUCUGCUUAGCCAGUGAACUGUUCUGAACUGCUAAUUGAUAAUAUGUAUGCGGCAACACCU---ACAUACAUACAUAUGUAUGUGUGUGUAUACUUA---U-------UU .....((((..((((((.(((....)))..))))...))..))))..((((.(((((((....)....---((((((((((....)))))))))))))))).))---)-------). ( -29.00) >DroEre_CAF1 5881 109 + 1 AUCAGCGAUCUGCUUAGUCAGUGAACUGUUCUGAACUGCUAAUUGAUAAUAUGUAUGGGGCAACACCUUGUACAUAUGUACAUAUG--------CAUAUACUUUUGAUUCGCACACU ....((((((...((((.(((....)))..))))..(((....((...(((((((..(((.....)))..)))))))...))...)--------)).........)).))))..... ( -28.70) >consensus AUCAGCGAUCUGCUUAGCCAGUGAACUGUUCUGAACUGCUAAUUGAUAAUAUGUAUGCGGCAACACCU___ACAUACAUACAUAUGUAUGUGUGUAUAUACUUA___U_______UU ((((((.....))((((.((((............)))))))).)))).....(((((((....).......((((((((((....))))))))))))))))................ (-15.44 = -18.12 + 2.69)

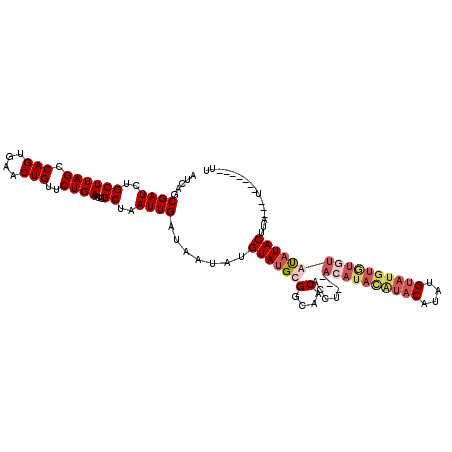
| Location | 4,651,345 – 4,651,445 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -13.61 |
| Energy contribution | -13.55 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4651345 100 - 20766785 UAAGUGGCAAAUGCGACGCAGUUUCAACUCGAAUCUAA-------A---UAUGUAAGUAUACACAUACGUAUGUACAUAUGU---AGGUGUUGCCGCAUACAUAUUAUCAAUU ...(((((((......((.(((....))))).(((((.-------(---((((((..(((((......)))))))))))).)---)))).)))))))................ ( -25.10) >DroSec_CAF1 5743 100 - 1 UAAGUGGCAAAUGCGACGCAGUUUCAACUCGAAUCUAA-------A---UAAGUAUACACAUACAUACAUAUGUAUGUAUGU---AGGUGUUGCCGCAUACAUAUUAUCAAUU ...(((((((((((.....((.(((.....))).))..-------.---...)))).(((.(((((((((....))))))))---).))))))))))................ ( -26.70) >DroSim_CAF1 6028 100 - 1 UAAGUGGCAAAUGCGACGCAGUUUCAACUCGAAUCUAA-------A---UAAGUAUACACACACAUACAUAUGUAUGUAUGU---AGGUGUUGCCGCAUACAUAUUAUCAAUU ...(((((((((((.....((.(((.....))).))..-------.---...)))).(((..((((((((....))))))))---..))))))))))................ ( -25.50) >DroEre_CAF1 5921 105 - 1 UAAGUAGAAAGUGCGACGCAGUUUUAGCUCGAAUCGAGUGUGCGAAUCAAAAGUAUAUG--------CAUAUGUACAUAUGUACAAGGUGUUGCCCCAUACAUAUUAUCAAUU ...(((....(.(((((((.......(((((...)))))(((((.(((....).)).))--------))).(((((....)))))..))))))))...)))............ ( -22.90) >consensus UAAGUGGCAAAUGCGACGCAGUUUCAACUCGAAUCUAA_______A___UAAGUAUACACACACAUACAUAUGUACAUAUGU___AGGUGUUGCCGCAUACAUAUUAUCAAUU ...(((((....(((((((((.(((.....))).))..............................((((((....)))))).....))))))).)).)))............ (-13.61 = -13.55 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:17 2006