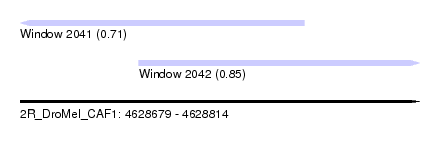
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,628,679 – 4,628,814 |
| Length | 135 |
| Max. P | 0.852667 |

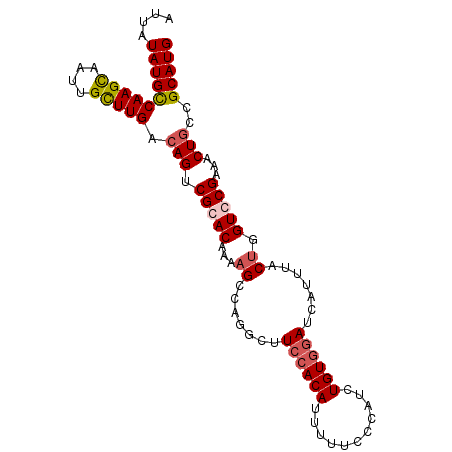
| Location | 4,628,679 – 4,628,775 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 88.37 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -17.93 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4628679 96 - 20766785 AUUAUAUGCCAAGUGAUUGCUUGACAGUCGGACAAAAGCCUGGCUUCCACAUUUUUCCCAUCUGUGGAUAAUUUACUGGUCCGAAACUGCCGCAUG ....((((((((((....))))).(((((((((...((.......((((((...........)))))).......)).))))))..)))..))))) ( -26.34) >DroSec_CAF1 5210 96 - 1 AUUAUAUGCCAAGCAAUUGCUUGACAGGCGCACAAAAGCCAGGCUUCCACAUUUUUCCCAUCUGUGGAUCAUUUACUGGUCCGAAACUGCCGCAUG ....((((((((((....)))))...((((.......(((((...((((((...........)))))).......))))).......))))))))) ( -28.14) >DroSim_CAF1 5212 96 - 1 AUUAUAUGCCAAGCAAUUGCUUGACAGCCGCACAAAAGCCAGGCUUCCACAUUUUUCCCAUCUGUGGAUCAUUUACUGGUCCGAAACUGCCGCAUG ......((.(((((....))))).))((.(((.....(((((...((((((...........)))))).......))))).......))).))... ( -24.70) >DroYak_CAF1 5254 96 - 1 AGAUUAUGUCAAGCAAUGGUUUGACAGUCGGACAAAAGCCUUCGUUCCACAUUUUUCUCAUCUGUCAAACAUUUACAGGUCCGAAACUCCCGCAUG ......((((((((....)))))))).((((((...(((....)))...............((((.........))))))))))............ ( -19.20) >consensus AUUAUAUGCCAAGCAAUUGCUUGACAGUCGCACAAAAGCCAGGCUUCCACAUUUUUCCCAUCUGUGGAUCAUUUACUGGUCCGAAACUGCCGCAUG ....((((((((((....))))).(((.(((((...((.......((((((...........)))))).......)).)))))...)))..))))) (-17.93 = -18.87 + 0.94)

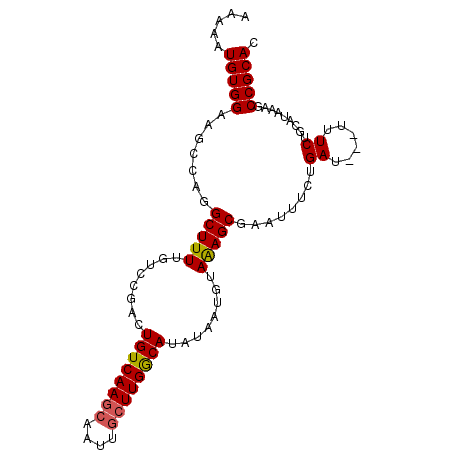
| Location | 4,628,719 – 4,628,814 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -18.75 |
| Energy contribution | -18.81 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4628719 95 + 20766785 AAAAAUGUGGAAGCCAGGCUUUUGUCCGACUGUCAAGCAAUCACUUGGCAUAUAAUGUAGAGCGAAUUUAUGAU-UUUUUCUGCAUAAAGCCGCAC .....(((((..((((((...((((..(.....)..))))...)))))).....((((((((.(((((...)))-)).))))))))....))))). ( -26.80) >DroSec_CAF1 5250 91 + 1 AAAAAUGUGGAAGCCUGGCUUUUGUGCGCCUGUCAAGCAAUUGCUUGGCAUAUAAUGUAAAGCGAAUUUCUGAU-----UCUGCAUAAAGCCGCAC ............((..((((((.((((...((((((((....)))))))).............(((((...)))-----)).)))))))))))).. ( -26.30) >DroSim_CAF1 5252 96 + 1 AAAAAUGUGGAAGCCUGGCUUUUGUGCGGCUGUCAAGCAAUUGCUUGGCAUAUAAUGUAAAGCGAAUUUCUGAUAUUUUUCAGCAUAAAGCCGCAC ........((((((...))))))(((((((((((((((....)))))))....................((((......)))).....)))))))) ( -30.90) >DroYak_CAF1 5294 94 + 1 AAAAAUGUGGAACGAAGGCUUUUGUCCGACUGUCAAACCAUUGCUUGACAUAAUCUGUAGAGCGAUUAUCUGAUG--UUUCUGUAUAAAGCCGCAC ................((((((...(.((..((((....(((((((.(((.....))).)))))))....)))).--..)).)...)))))).... ( -19.60) >consensus AAAAAUGUGGAAGCCAGGCUUUUGUCCGACUGUCAAGCAAUUGCUUGGCAUAUAAUGUAAAGCGAAUUUCUGAU___UUUCUGCAUAAAGCCGCAC .....(((((.......(((((........((((((((....))))))))........)))))........((......)).........))))). (-18.75 = -18.81 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:03 2006