
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,625,496 – 4,625,716 |
| Length | 220 |
| Max. P | 0.589342 |

| Location | 4,625,496 – 4,625,616 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -33.99 |
| Consensus MFE | -20.17 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
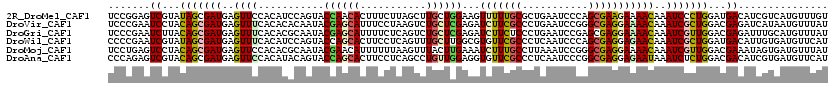
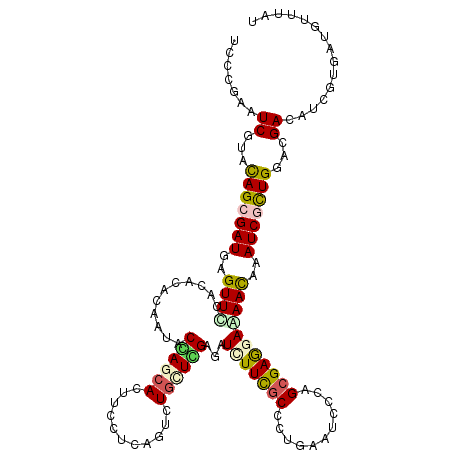
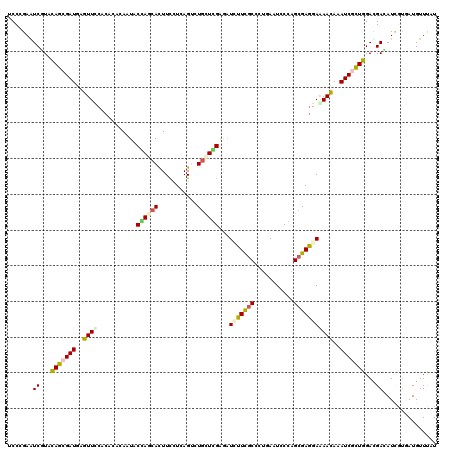
>2R_DroMel_CAF1 4625496 120 - 20766785 UCCGGAGUCGUAUAGCGAUGAGUUCCACAUCCAGUACCAACACUUUCUUAGCUUGCUGGAAGUUUUUGCGCUGAAUCCCAGCGAAGAAAACAAAUCCCUGGAUGACAUCGUCAUGUUUGU ..........(((..(((((...((((..(((((((.................))))))).((((((.(((((.....)))))..)))))).......))))...)))))..)))..... ( -31.33) >DroVir_CAF1 2094 120 - 1 UCCCGAAUCCUACAGCGAUGAGUUUCACACACAAUACGAGCAUUUCCUAAGUCUGCUCGAGAUCUUCGCCCUGAAUCCGGGCGAGGAAAACAAAUCGCUGGACGAGAUCAUAAUGUUUAU ....((.((...(((((((..((((...........((((((...........))))))...(((((((((.......)))))))))))))..)))))))...))..))........... ( -38.90) >DroGri_CAF1 2150 120 - 1 UCCCGAAUCUUACAGCGAUGAGUUUCACACGCAAUACGAGCAUUUUCUCAGUCUGCUCGAGAUCUUCUCCCUGAAUCCGAGCGAGGAAAACAAAUCGUUGGACGAGAUUUGCAUGUUUAU ...((((((((.(((((((..((((..(.(((.....(((((...........)))))(((.....)))...........))).)..))))..)))))))...))))))))......... ( -33.80) >DroWil_CAF1 2887 120 - 1 CCCCGAAUCGUAUAGCGAUGAGUUUCACAUCCAGUACCAGCACUUCCUCAGUUUGCUUGGCGUGUUCGCCCUCAAUCCCAGCGAGGAGAACAAAUCGCUGGAUGACAUUGUGAUGUUCAU ....(((.(((...((((((.(...).((((((((.((((((...........))).)))..(((((..((((.........)))).)))))....)))))))).)))))).)))))).. ( -33.40) >DroMoj_CAF1 2242 120 - 1 UCCUGAGUCCUACAGCGAUGAGUUCCACACGCAAUACGAACAUUUUUUAAGUUUACUUGAAAUCUUUGCCUUAAAUCCGGGCGAGGAAAACAAAUCGUUGGACGAAAUAGUGAUGUUUAU ......((((....(((.((.......)))))...((((.....(((((((....)))))))(((((((((.......))))))))).......)))).)))).(((((....))))).. ( -29.50) >DroAna_CAF1 2020 120 - 1 CCCAGAGUCGUACAGCGAUGAGUUCCACAUACAGUACCAGCACUUCCUCAGCCUGUUGGAGGUGUUCGCCCUCAAUCCCGGCGAGGAGAAUAAAUCUCUGGACGACAUCGUGAUGUUCAU ....(((((((.(((.(((..((((........((....))...(((((.(((.((((..(((....)))..))))...))))))))))))..))).))).))))).))........... ( -37.00) >consensus UCCCGAAUCGUACAGCGAUGAGUUCCACACACAAUACCAGCACUUCCUCAGUCUGCUCGAGAUCUUCGCCCUGAAUCCCAGCGAGGAAAACAAAUCGCUGGACGACAUCGUGAUGUUUAU .......((...(((((((..((((...........((((((...........))))))...(((((((...........)))))))))))..)))))))...))............... (-20.17 = -20.32 + 0.14)
| Location | 4,625,616 – 4,625,716 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
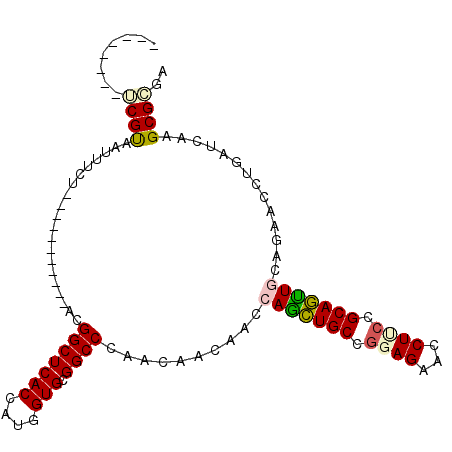
>2R_DroMel_CAF1 4625616 100 - 20766785 --------UCGUAGUUUUUAUACUGUUAUAACGGCUCACCAUGGUGCGGCCCAACAACAACCAGCUGCCGGAGAACCUUCCGCAGUUGCAGAACCUCAUCAAGCGGGA --------..............(((((.....(((((((....))).))))..........(((((((.((((....))))))))))).............))))).. ( -26.80) >DroPse_CAF1 2173 89 - 1 --------UCGCAAUUUCU-----------UCGGCUCACCAUGGUGCGGCCCAACAACAAUCAACUGCCGGAGAAUCUUCCGCAGUUACAGAACCUGAUCAAGCGCGA --------((((.......-----------..(((((((....))).))))...........((((((.((((....)))))))))).................)))) ( -25.30) >DroEre_CAF1 2130 100 - 1 --------UCGUAGUUUUUAUAUUCUUAAAACGGCUCACUAUGGUGCGGCCCAACAACAACCAGUUGCCGGAGAACCUGCCGCAAUUGCAGAACCUGAUCAAGCGGGA --------..((.(((((.........)))))(((((((....))).))))..))......(((((((.((........))))))))).....((((......)))). ( -22.50) >DroYak_CAF1 2136 100 - 1 --------UCGUAGUUAUUAUCCGUUAAGAACGGCUCACCAUGGUGCGGCCUAACAACAACCAGCUGCCGGAGAACCUCCCGCAGUUGCAGAACCUAAUCAAGCGGGA --------............((((((......(((((((....))).))))..........(((((((.((((...)))).))))))).............)))))). ( -31.60) >DroAna_CAF1 2140 97 - 1 GAAAUAAGGCGUAAUUUCC-----------CCGGCUCACGAUGGUGCGGCCUAACAAUAAUCAGCUGCCAGAGAACCUGCCGCAGCUGCAGAACCUGAUCAAGCGCGA ........((((.......-----------..(((((((....))).))))..........((((((((((.....)))..)))))))..............)))).. ( -27.20) >DroPer_CAF1 1993 89 - 1 --------UCGCAAUUUCU-----------UCGGCUCACCAUGGUGCGGCCCAACAACAAUCAACUGCCGGAGAAUCUUCCGCAGUUACAGAACCUGAUCAAGCGCGA --------((((.......-----------..(((((((....))).))))...........((((((.((((....)))))))))).................)))) ( -25.30) >consensus ________UCGUAAUUUCU___________ACGGCUCACCAUGGUGCGGCCCAACAACAACCAGCUGCCGGAGAACCUUCCGCAGUUGCAGAACCUGAUCAAGCGCGA ........((((....................(((((((....))).))))..........(((((((.((((...)))).)))))))..............)))).. (-22.64 = -22.70 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:00 2006