| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,613,329 – 4,613,449 |
| Length | 120 |
| Max. P | 0.823492 |

| Location | 4,613,329 – 4,613,449 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.89 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -22.07 |
| Energy contribution | -23.52 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4613329 120 + 20766785 UGAACUCCGAAAAUGAGAACUUGCCAUUGGCACAGCACUGUUUUAUUCAGGCUGUAGUGCUGGAAAAAAAGUGCUACACAGCGUGGACCAAUCUCGGAGUUCUCUAUAUUAAACUGAAUG .(((((((((.....((..((((((...)))).))..))......((((.(((((((..((........))..))..))))).))))......))))))))).................. ( -36.00) >DroVir_CAF1 3408 120 + 1 UGCACGCCGCCAACGAGAAUCUGCCUGUGGCGCAGCAUUGUUUCAUUCAGGCAGUUGUGCUGGAUCGAAAAUCCUACACGGCCUGGACAAAUCUGGGUGUGCUGUACAUCAAGCUGAACG .((((((((((.........((((((((((.((......)).)))..)))))))..(((..((((.....))))..))))))..(((....))).))))))).................. ( -38.20) >DroGri_CAF1 3373 120 + 1 UGUACUCCGCCAGCGAGAAUCUGCCCAUGGCGCAGCAUUGUUUAAUUCAGGCACUGAUGCUGGACCGCAAAUCCUACACAGCAUGGACGAACCUGGGUGUGCUGUACAUAAAGCUGGGCA ........((((((........(((...)))(((((((......(((((((...(.((((((................)))))).).....)))))))))))))).......))).))). ( -34.69) >DroEre_CAF1 3111 120 + 1 UGCACUCCGAAUAUGAAAACUUGUCAUUGGCACAGCACUGUUUUAUUCAAGCUGUGGAGCUCGAAAGAAAGUGCUACACAGCGUGGACCAAUCUCGGAGUGCUCUAUAUUAAACUAAAUG .(((((((((...(((.......)))((((.(((....)))....((((.((((((.(((((....))....))).)))))).))))))))..))))))))).................. ( -37.70) >DroYak_CAF1 3064 120 + 1 UGCACUCCGAAUAUGAGAACUUGCCAUUGGCCCAGCACUGUUUUAUUCAGGCUGUGGAACUAGAAAGAAAGUGCUACACAGCGUGGACCAAUCUCGGAGUGCUUUAUAUUAAACUAAAAG .(((((((((.....(((((.(((..........)))..))))).((((.((((((..(((........)))....)))))).))))......))))))))).................. ( -36.60) >DroAna_CAF1 3084 120 + 1 UGCACUCGGAUCACGAAAACUUGCCGUUGGCCCAGCACUGUUUCAUACAGGCCGUGGAGCUGGAACGAAAGUGUUUCACGGCCUGGACGAACCUCGGGGUUCUGUACGUAAAAAUAGACA (((..(((.....)))......(((...)))...)))((((((....(((((((((((((....((....)))))))))))))))...(((((....))))).........))))))... ( -42.60) >consensus UGCACUCCGAAAACGAGAACUUGCCAUUGGCACAGCACUGUUUUAUUCAGGCUGUGGAGCUGGAAAGAAAGUGCUACACAGCGUGGACCAAUCUCGGAGUGCUGUACAUUAAACUAAACG .(((((((((............(((...)))..............((((.((((((..(((........)))....)))))).))))......))))))))).................. (-22.07 = -23.52 + 1.45)

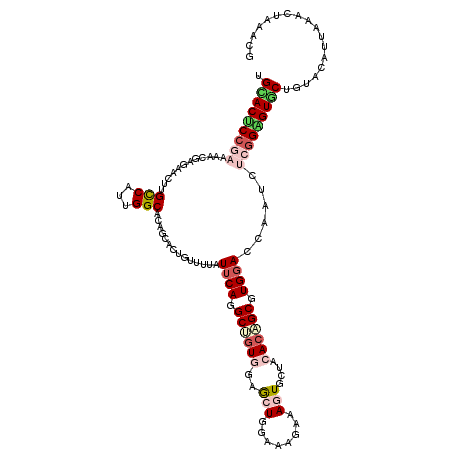
| Location | 4,613,329 – 4,613,449 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.89 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4613329 120 - 20766785 CAUUCAGUUUAAUAUAGAGAACUCCGAGAUUGGUCCACGCUGUGUAGCACUUUUUUUCCAGCACUACAGCCUGAAUAAAACAGUGCUGUGCCAAUGGCAAGUUCUCAUUUUCGGAGUUCA ..................(((((((((((.(((..(..((((..(((((((.((((..((((......).)))...)))).)))))))..)....)))..)..).)).))))))))))). ( -37.80) >DroVir_CAF1 3408 120 - 1 CGUUCAGCUUGAUGUACAGCACACCCAGAUUUGUCCAGGCCGUGUAGGAUUUUCGAUCCAGCACAACUGCCUGAAUGAAACAAUGCUGCGCCACAGGCAGAUUCUCGUUGGCGGCGUGCA .((.((......)).)).((((.....(.((..(.(((((.((((.(((((...))))).))))....))))).)..)).)...((((((((...))).(.....)....))))))))). ( -36.70) >DroGri_CAF1 3373 120 - 1 UGCCCAGCUUUAUGUACAGCACACCCAGGUUCGUCCAUGCUGUGUAGGAUUUGCGGUCCAGCAUCAGUGCCUGAAUUAAACAAUGCUGCGCCAUGGGCAGAUUCUCGCUGGCGGAGUACA .(((.(((......(((((((......((.....)).))))))).((((((((((((.((((((..((...........)).)))))).)))....))))))))).))))))........ ( -35.30) >DroEre_CAF1 3111 120 - 1 CAUUUAGUUUAAUAUAGAGCACUCCGAGAUUGGUCCACGCUGUGUAGCACUUUCUUUCGAGCUCCACAGCUUGAAUAAAACAGUGCUGUGCCAAUGACAAGUUUUCAUAUUCGGAGUGCA ..................((((((((((.............(..(((((((((..((((((((....))))))))..))..)))))))..)..((((.......)))).)))))))))). ( -40.40) >DroYak_CAF1 3064 120 - 1 CUUUUAGUUUAAUAUAAAGCACUCCGAGAUUGGUCCACGCUGUGUAGCACUUUCUUUCUAGUUCCACAGCCUGAAUAAAACAGUGCUGGGCCAAUGGCAAGUUCUCAUAUUCGGAGUGCA ..................((((((((((((((((((((((((((.(((............))).))))))(((.......))).).))))))))).....(....)...)))))))))). ( -39.60) >DroAna_CAF1 3084 120 - 1 UGUCUAUUUUUACGUACAGAACCCCGAGGUUCGUCCAGGCCGUGAAACACUUUCGUUCCAGCUCCACGGCCUGUAUGAAACAGUGCUGGGCCAACGGCAAGUUUUCGUGAUCCGAGUGCA .............((((.(((((....)))))...(((((((((....................)))))))))(((((((...(((((......)))))...)))))))......)))). ( -38.95) >consensus CGUUCAGUUUAAUAUACAGCACUCCGAGAUUGGUCCACGCUGUGUAGCACUUUCGUUCCAGCACCACAGCCUGAAUAAAACAGUGCUGCGCCAAUGGCAAGUUCUCAUUUUCGGAGUGCA ..................((((.((((((((.(((......((((((((((....(((..(((....)))..)))......))))))))))....))).)))).......)))).)))). (-17.06 = -17.84 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:56 2006