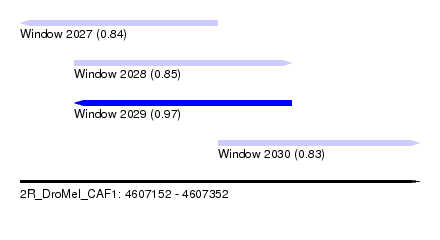
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,607,152 – 4,607,352 |
| Length | 200 |
| Max. P | 0.965637 |

| Location | 4,607,152 – 4,607,251 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -15.26 |
| Energy contribution | -16.78 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
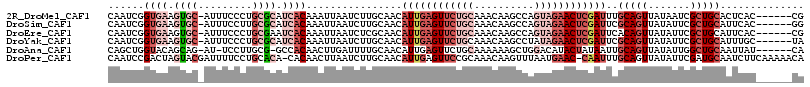
>2R_DroMel_CAF1 4607152 99 - 20766785 AAUCUUGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUUGCAGUUAUAAUCGCUGCACUCACCGGUCGCCUAUCAUAUCU---AUAAUACUUAC ......((.((((((((((((((..........)))))))))))).((((((.......)))))).......)).))...........---........... ( -23.30) >DroSec_CAF1 7427 99 - 1 AAUCUUGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUCGCAGUUAUAUUCGCUGCAUUCACGGGUCGCCUACAUUAUUU---AUAAUACUCAC ......((.((((((((((((((..........))))))))))))..(((((.......)))))........)).))...........---........... ( -23.00) >DroSim_CAF1 7830 99 - 1 AAUCUUGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUCGCAGUUAUAUUCGCUGCAUUCACGGGUCGCCUACCAUAUCU---AUAAUACUCAC ......((.((((((((((((((..........))))))))))))..(((((.......)))))........)).))...........---........... ( -23.00) >DroEre_CAF1 7191 99 - 1 AAUCUCGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUCACAGUUAUAUUCGCUGCAUUCACCGGUCGCCUAUCAUAUCU---GAAUUACUUAC ...........((((((((((((..........))))))))))))...(((.(((((..((.((........)).))..)).))).))---).......... ( -19.30) >DroYak_CAF1 7571 99 - 1 AAUCUUGCAACAUUGAGUUCUGCAAACAAGCCUAUAGAACUCGAUUCGCAGUUAUAUUCGCUGCAUUUGCUAACUGCCUAACAUAUCU---CAAUUACUUAC ......((((.((((((((((((......))....))))))))))..(((((.......)))))..))))..................---........... ( -20.70) >DroAna_CAF1 6465 102 - 1 GAUUUUGCAACAUUGAGUUCUGCAAAAAAGCUGGACAUACUAUAAUUGCAGUUAUAUUGGCUGCAAUUAUCAGAACUUUAGGAACUUUGAGAUAAUACUUAC ..............(((((((((......))........((((((((((((((.....)))))))))))).)).......)))))))((((......)))). ( -21.80) >consensus AAUCUUGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUCGCAGUUAUAUUCGCUGCAUUCACCGGUCGCCUACCAUAUCU___AUAAUACUUAC ......((...((((((((((((..........))))))))))))..(((((.......)))))...........))......................... (-15.26 = -16.78 + 1.53)


| Location | 4,607,179 – 4,607,288 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -19.48 |
| Energy contribution | -18.96 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4607179 109 + 20766785 CG------GUGAGUGCAGCGAUUAUAACUGCAAAUCGAGUUCUACUGGCUUGUUUGCAGAACUCAAUGUUGCAAGAUUAAUUUGUGAUGCGCAGGGAAAU-GCACUUCACCGAUUG ((------((((((((((((((((((((((((((.((((((.....)))))))))))))(((.....)))...........))))))).))).......)-)))).)))))).... ( -37.01) >DroSim_CAF1 7857 109 + 1 CC------GUGAAUGCAGCGAAUAUAACUGCGAAUCGAGUUCUACUGGCUUGUUUGCAGAACUCAAUGUUGCAAGAUUAAUUUGUGAUGCGCAAGGAAAU-GCACUUCACCGAUUG ..------.....(((((((.......(((((((.((((((.....))))))))))))).......)))))))....(((((.((((.(.(((......)-)).).)))).))))) ( -30.24) >DroEre_CAF1 7218 109 + 1 CG------GUGAAUGCAGCGAAUAUAACUGUGAAUCGAGUUCUACUGGCUUGUUUGCAGAACUCAAUGUUGCGAGAUUAAUUUGUGAUUCGCAGGGAAAU-GCACUUCACCGAUUG ((------(((((((((..........((((((((((((((((....((......)))))))))......(((((.....)))))))))))))).....)-))).))))))).... ( -37.76) >DroYak_CAF1 7598 109 + 1 UA------GCAAAUGCAGCGAAUAUAACUGCGAAUCGAGUUCUAUAGGCUUGUUUGCAGAACUCAAUGUUGCAAGAUUAAUUUGUGAUGCGCAGGGAAAU-GCACUUCACCGAUUG ..------.....(((((((.......(((((((.((((((.....))))))))))))).......)))))))....(((((.((((.(.(((......)-)).).)))).))))) ( -31.24) >DroAna_CAF1 6495 107 + 1 UG------AUAAUUGCAGCCAAUAUAACUGCAAUUAUAGUAUGUCCAGCUUUUUUGCAGAACUCAAUGUUGCAAAAUCAAGUUGUGGC-CGCAAGGA-AU-CUGCUGUACCAGCUG ..------((((((((((.........))))))))))........(((((.....(((((.......((..(((.......)))..))-(....)..-.)-))))......))))) ( -29.00) >DroPer_CAF1 7072 114 + 1 UGUUUUUGAAGAUUGCAUCGAAUAUAACUGCAAAUUG-GUUCAUUAAACUUGUUUGCGGAACUCAAUGUUGCAAGAUUAAGUUGUG-UGUGCAGGAAAAUCGUACUAGUCGGAUUG .((...(((...((((((...((((((((((((((.(-(((.....)))).))))))(.(((.....))).).......)))))))-))))))).....))).))........... ( -23.20) >consensus CG______GUGAAUGCAGCGAAUAUAACUGCAAAUCGAGUUCUACUGGCUUGUUUGCAGAACUCAAUGUUGCAAGAUUAAUUUGUGAUGCGCAGGGAAAU_GCACUUCACCGAUUG .............(((((((.......(((((((.((((((.....))))))))))))).......)))))))........((((.....))))...................... (-19.48 = -18.96 + -0.52)
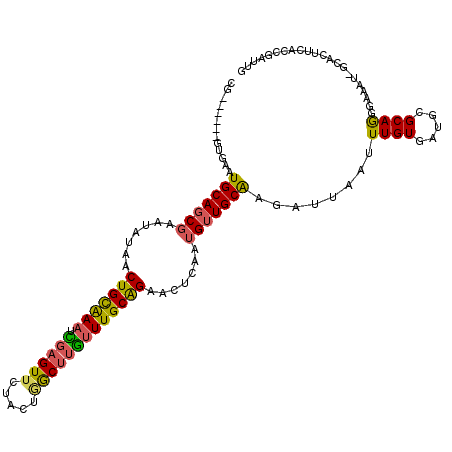
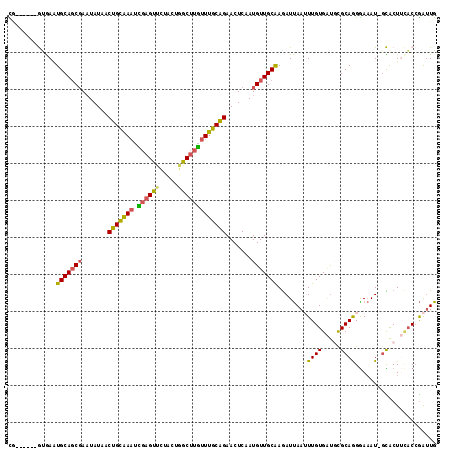
| Location | 4,607,179 – 4,607,288 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -13.90 |
| Energy contribution | -17.52 |
| Covariance contribution | 3.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
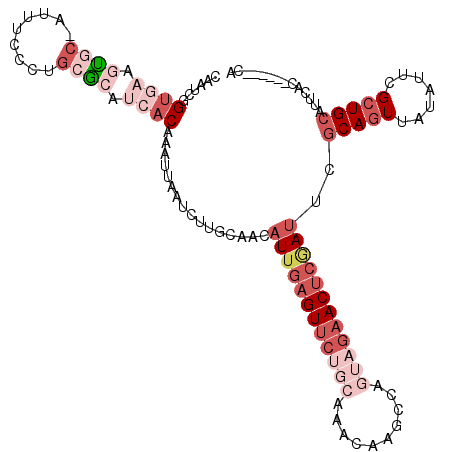
>2R_DroMel_CAF1 4607179 109 - 20766785 CAAUCGGUGAAGUGC-AUUUCCCUGCGCAUCACAAAUUAAUCUUGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUUGCAGUUAUAAUCGCUGCACUCAC------CG ....((((((.((((-(.......(((........((((...((((((.((((((((((((..........))))))))))))))))))...))))))))))))))))------)) ( -36.61) >DroSim_CAF1 7857 109 - 1 CAAUCGGUGAAGUGC-AUUUCCUUGCGCAUCACAAAUUAAUCUUGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUCGCAGUUAUAUUCGCUGCAUUCAC------GG ....((((((.((((-(......))))).))))................((((((((((((..........))))))))))))..(((((.......))))).....)------). ( -33.60) >DroEre_CAF1 7218 109 - 1 CAAUCGGUGAAGUGC-AUUUCCCUGCGAAUCACAAAUUAAUCUCGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUCACAGUUAUAUUCGCUGCAUUCAC------CG ....(((((((.(((-(......(((((..............)))))..((((((((((((..........))))))))))))................)))))))))------)) ( -32.54) >DroYak_CAF1 7598 109 - 1 CAAUCGGUGAAGUGC-AUUUCCCUGCGCAUCACAAAUUAAUCUUGCAACAUUGAGUUCUGCAAACAAGCCUAUAGAACUCGAUUCGCAGUUAUAUUCGCUGCAUUUGC------UA .(((..((((.((((-(......))))).))))..)))......((((.((((((((((((......))....))))))))))..(((((.......)))))..))))------.. ( -32.50) >DroAna_CAF1 6495 107 - 1 CAGCUGGUACAGCAG-AU-UCCUUGCG-GCCACAACUUGAUUUUGCAACAUUGAGUUCUGCAAAAAAGCUGGACAUACUAUAAUUGCAGUUAUAUUGGCUGCAAUUAU------CA ((((((((...((((-..-...)))).-))).........(((((((((.....))..))))))).)))))........((((((((((((.....))))))))))))------.. ( -32.10) >DroPer_CAF1 7072 114 - 1 CAAUCCGACUAGUACGAUUUUCCUGCACA-CACAACUUAAUCUUGCAACAUUGAGUUCCGCAAACAAGUUUAAUGAAC-CAAUUUGCAGUUAUAUUCGAUGCAAUCUUCAAAAACA ...........((((((..(..(((((..-...((((((((........))))))))..........((((...))))-.....)))))..)...))).))).............. ( -14.70) >consensus CAAUCGGUGAAGUGC_AUUUCCCUGCGCAUCACAAAUUAAUCUUGCAACAUUGAGUUCUGCAAACAAGCCAGUAGAACUCGAUUCGCAGUUAUAUUCGCUGCAUUCAC______CA ......((((.((((.........)))).))))................((((((((((((..........))))))))))))..(((((.......))))).............. (-13.90 = -17.52 + 3.61)
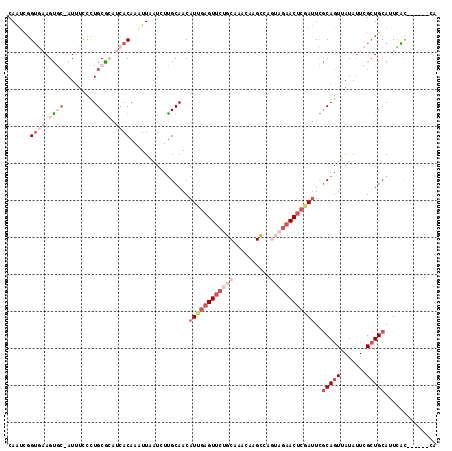
| Location | 4,607,251 – 4,607,352 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -19.51 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4607251 101 + 20766785 AAUUUGUGAUGCGCAGGGAAAU-GCACUUCACCGAUUGCAAUUCAAAUUGCAGUCACGUGGUCGAGAUUCCAUUGCAGUCUCAACAAA---UUAA-AAAUUAAAAA (((((((((.((((((((((..-.(...((((.(((((((((....)))))))))..))))..)...)))).)))).)).)).)))))---))..-.......... ( -27.80) >DroSec_CAF1 7526 101 + 1 AAUUUGUGAUGCGCAAGGAAAU-GCACUUCACCGAUUGCAACUCAAAUUGCAGUCACGUGGUCAAGAAUCCAUUGCAGUCUCAACAAA---UUAA-CAAUUAAAAA (((((((((.(((((((((...-.....((((.((((((((......))))))))..)))).......))).)))).)).)).)))))---))..-.......... ( -25.86) >DroSim_CAF1 7929 101 + 1 AAUUUGUGAUGCGCAAGGAAAU-GCACUUCACCGAUUGCAACUCAAAUUGCAGUCACGUGGUCGAGAAUCCAUUGCAGUCUCAACAAA---UUAA-CAAUUAAAAA (((((((((.(((((((((...-.....((((.((((((((......))))))))..)))).......))).)))).)).)).)))))---))..-.......... ( -25.86) >DroEre_CAF1 7290 102 + 1 AAUUUGUGAUUCGCAGGGAAAU-GCACUUCACCGAUUGCACUUCAAAUUGCAGUCACGUGGUUGAGAAUCCAUUGCAAUCUCAACAAA---UUAUCCAAAUUAAUG ((((((.(((.(((.(((((..-....))).))(((((((........)))))))..)))(((((((...........)))))))...---..))))))))).... ( -26.20) >DroYak_CAF1 7670 101 + 1 AAUUUGUGAUGCGCAGGGAAAU-GCACUUCACCGAUUGCAACUGAAGUUGCAGUCACGUGGUCGAGAAUCCAUUGCAGUCUCAACAAA---UUAA-CAACUUAAUA (((((((((.(((((((((...-.....((((.(((((((((....)))))))))..)))).......))).)))).)).)).)))))---))..-.......... ( -29.16) >DroPer_CAF1 7149 103 + 1 AAGUUGUG-UGUGCAGGAAAAUCGUACUAGUCGGAUUGCAAUGCAUUUUGCAGUCACGUGGUCUAGAACGCUCUGCUGCUUAAACAGCCCCUUUG-CCAUUAGAA- ..(((((.-...((((.(....(((.((((.((((((((((......)))))))).))....)))).)))...).))))....))))).......-.........- ( -30.00) >consensus AAUUUGUGAUGCGCAGGGAAAU_GCACUUCACCGAUUGCAACUCAAAUUGCAGUCACGUGGUCGAGAAUCCAUUGCAGUCUCAACAAA___UUAA_CAAUUAAAAA ..(((((((.(((((((((......(((.....((((((((......))))))))....)))......))).)))).)).)).))))).................. (-19.51 = -19.68 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:51 2006