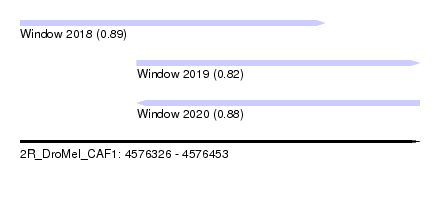
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,576,326 – 4,576,453 |
| Length | 127 |
| Max. P | 0.893155 |

| Location | 4,576,326 – 4,576,423 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.16 |
| Mean single sequence MFE | -17.10 |
| Consensus MFE | -14.17 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4576326 97 + 20766785 UCCCCAUUCUCCAUCUGCAUCAUCCAUCCUCCGGCGAACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACACGGCUGCAAUCAGCAGCAUCCACA ................................((...........(((((((....)))))))............(((((.....)))))..))... ( -15.20) >DroSec_CAF1 7771 97 + 1 UCCCCAUUCCCCAUCUGCAUCAUCCAUCCUCCGGCGAACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACGCGGCUGCAAUCAGCAGCAUUCACA ...............(((.((..............(((....)))(((((((....)))))))......)).)))(((((.....)))))....... ( -17.60) >DroSim_CAF1 8068 97 + 1 UCCCCAUUCCCCAUCUGCAUCAUCCAUCCUCCGGCGAACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACGCGGCUGCAAUCAGCAGCAUUCACA ...............(((.((..............(((....)))(((((((....)))))))......)).)))(((((.....)))))....... ( -17.60) >DroEre_CAF1 14706 96 + 1 UUCCCAUUGCCCAUCUCCAUUACCCAUCCUCCGGCGAACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACGCGGCUG-CAUCAGCGGCAUCCACA ......(((((.....................)))))........(((((((....)))))))......((.((.((((-...)))).)).)).... ( -18.00) >consensus UCCCCAUUCCCCAUCUGCAUCAUCCAUCCUCCGGCGAACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACGCGGCUGCAAUCAGCAGCAUCCACA .................................(((.........(((((((....)))))))........))).(((((.....)))))....... (-14.17 = -14.48 + 0.31)

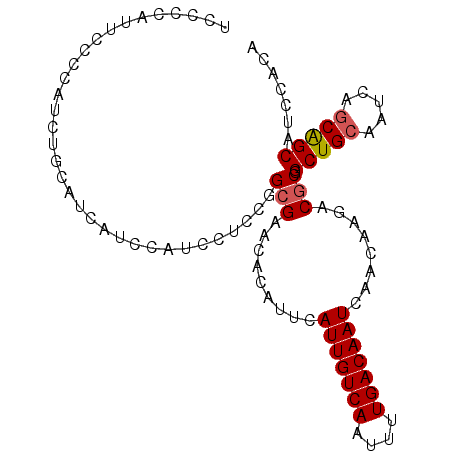
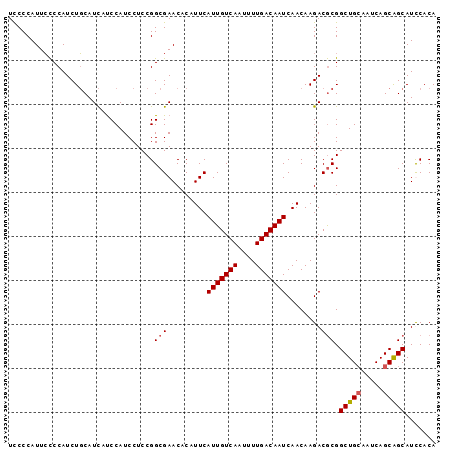
| Location | 4,576,363 – 4,576,453 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 87.37 |
| Mean single sequence MFE | -16.66 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.02 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4576363 90 + 20766785 ACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACACGGCUGCAAUCAGCAGCAUCCACAUA------CACGCGAAAAAAUAUAUAGAAAUAUGCU .....((((((((((....)))))))............(((((.....))))).........------.....)))......((((....)))).. ( -16.50) >DroSec_CAF1 7808 84 + 1 ACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACGCGGCUGCAAUCAGCAGCAUUCACAUA------CAGGCGAAAAAAUAG------AUAUGCU ...((((((((((((....)))))))........(((.(((((.....))))).....(...------..))))........)------).))).. ( -17.10) >DroSim_CAF1 8105 84 + 1 ACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACGCGGCUGCAAUCAGCAGCAUUCACAUA------CAGACGAAAAAAUAG------AUAUGCU .....((((((((((....)))))))............(((((.....))))).........------.....))).......------....... ( -15.30) >DroEre_CAF1 14743 88 + 1 ACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACGCGGCUG-CAUCAGCGGCAUCCACAUACGAGUACAGGCGAAG-AAUAG------UUAUGCU ....(((((((((((....)))))))........(((.((((-(....)))))....((......))....)))..)-)))..------....... ( -18.80) >DroYak_CAF1 14700 83 + 1 AGACAUUCAUUGUCAAUUUUGACAAUCAACAAGACACGGCUGCCAUCAGCAGCAUCCACAUA------CAGGCGAGA-AAUAG------AUAUGGU ...((((((((((((....)))))))..........(((((((.....)))))..((.....------..))))...-....)------).))).. ( -15.60) >consensus ACACAUUCAUUGUCAAUUUUGACAAUCAACAAGACGCGGCUGCAAUCAGCAGCAUCCACAUA______CAGGCGAAAAAAUAG______AUAUGCU .....((((((((((....)))))))............(((((.....)))))....................))).................... (-14.14 = -14.02 + -0.12)

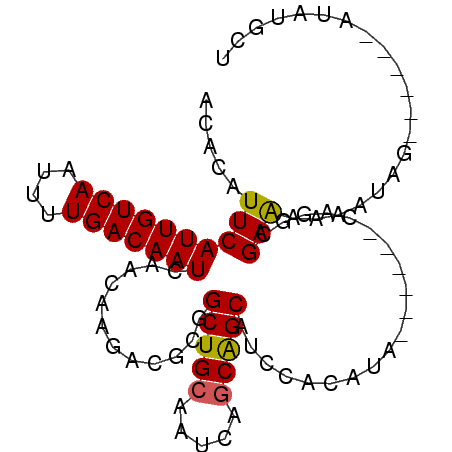
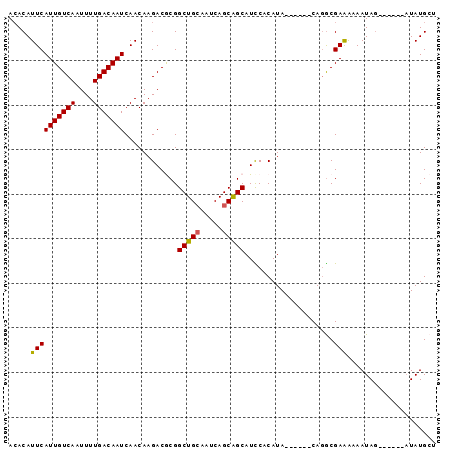
| Location | 4,576,363 – 4,576,453 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 87.37 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
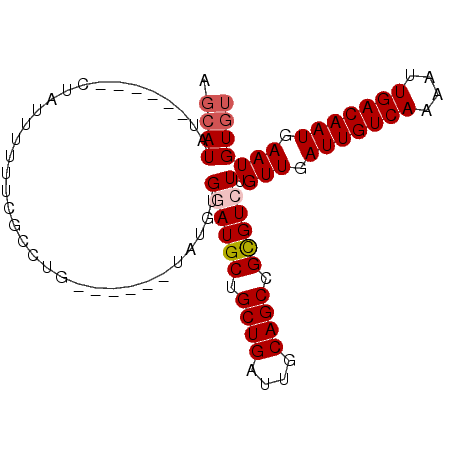
>2R_DroMel_CAF1 4576363 90 - 20766785 AGCAUAUUUCUAUAUAUUUUUUCGCGUG------UAUGUGGAUGCUGCUGAUUGCAGCCGUGUCUUGUUGAUUGUCAAAAUUGACAAUGAAUGUGU .((((...(((((((((..........)------)))))))).(((((.....))))).))))...(((.(((((((....))))))).))).... ( -24.90) >DroSec_CAF1 7808 84 - 1 AGCAUAU------CUAUUUUUUCGCCUG------UAUGUGAAUGCUGCUGAUUGCAGCCGCGUCUUGUUGAUUGUCAAAAUUGACAAUGAAUGUGU .((....------.......(((((...------...))))).(((((.....))))).)).....(((.(((((((....))))))).))).... ( -22.40) >DroSim_CAF1 8105 84 - 1 AGCAUAU------CUAUUUUUUCGUCUG------UAUGUGAAUGCUGCUGAUUGCAGCCGCGUCUUGUUGAUUGUCAAAAUUGACAAUGAAUGUGU .((((((------..((......))..)------)))))....(((((.....)))))((((((((((..(((.....)))..)))).).))))). ( -21.40) >DroEre_CAF1 14743 88 - 1 AGCAUAA------CUAUU-CUUCGCCUGUACUCGUAUGUGGAUGCCGCUGAUG-CAGCCGCGUCUUGUUGAUUGUCAAAAUUGACAAUGAAUGUGU .(((((.------.....-(.......)......)))))((((((.((((...-)))).)))))).(((.(((((((....))))))).))).... ( -22.60) >DroYak_CAF1 14700 83 - 1 ACCAUAU------CUAUU-UCUCGCCUG------UAUGUGGAUGCUGCUGAUGGCAGCCGUGUCUUGUUGAUUGUCAAAAUUGACAAUGAAUGUCU .((((((------(....-........)------.))))))..(((((.....)))))........(((.(((((((....))))))).))).... ( -21.00) >consensus AGCAUAU______CUAUUUUUUCGCCUG______UAUGUGGAUGCUGCUGAUUGCAGCCGCGUCUUGUUGAUUGUCAAAAUUGACAAUGAAUGUGU .((((..................................((((((.((((....)))).)))))).(((.(((((((....))))))).))))))) (-19.10 = -19.66 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:41 2006