
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
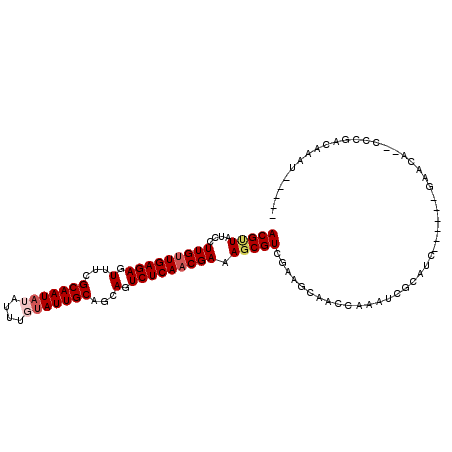
| Location | 4,571,667 – 4,571,782 |
| Length | 115 |
| Max. P | 0.644466 |

| Location | 4,571,667 – 4,571,763 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -18.45 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4571667 96 - 20766785 ACGGUAUCCUUGUUGAGAGUUUCGCAAUAUAUUUUCAUUGCUGCAGUCUCAUCGAAAGCGUCGAAGCAACCGAAUCGCAUC------GAACA--UCCGACAAAU----- .(((.........(((((.(...(((((........)))))...).)))))((((..((((((.......)))..))).))------))...--.)))......----- ( -20.10) >DroVir_CAF1 13231 107 - 1 ACGCUAUCCUUGUUGAGAGUUUCGCAAUAUAUUUGUAUUGCAGCAGUCUCAACGAAAGCGUCGAAGCAACCAAAUCGCAACCGAAUCGAAAA--CGCUCGAAAUCAAAC .((((....(((((((((.(...(((((((....)))))))...).))))))))).))))((((.((.......(((....)))...(....--)))))))........ ( -26.70) >DroPse_CAF1 8412 96 - 1 ACGUUAUCCUUGUUGAGAGUUUCGCAAUAUAUUUGUAUUGCAGCAGUCUCAACGAAAGCGUCGAACCAACCUAAUCGCAUC------GAACA--CCCGAUUAAA----- (((((....(((((((((.(...(((((((....)))))))...).))))))))).)))))..........((((((....------.....--..))))))..----- ( -22.80) >DroMoj_CAF1 15435 109 - 1 ACGUUAUCCUUGUUGAGAGUUUUGCAAUAUAUUUGUAUUGCAGCAGUCUCAACGAAAGCGUCGAAGCAACCAACUCGCAGCCCAAUCGAAAACACGCUCGAAACGUAAC (((((....(((((((((.(.(((((((((....))))))))).).)))))))))..((......)).......(((.(((..............)))))))))))... ( -30.54) >DroAna_CAF1 10697 102 - 1 ACGCUAUCCUUGUUGAGAGUUUCGCAAUAUAUUUUUAUUGCAGCAGUCUCAACGAAAGCGUCGAAGCAACCGAAUCGCAUCGAAAUCGAACA--CCCGACAAAU----- ..(((....(((((((((.(...((((((......))))))...).))))))))).)))((((..((.........)).(((....)))...--..))))....----- ( -27.90) >DroPer_CAF1 8525 96 - 1 ACGUUAUCCUUGUUGAGAGUUUCGCAAUAUAUUUGUAUUGCAGCAGUCUCAACGAAAGCGUCGAACCAACCUAAUCGCAUC------GAACA--CCCGAUUAAA----- (((((....(((((((((.(...(((((((....)))))))...).))))))))).)))))..........((((((....------.....--..))))))..----- ( -22.80) >consensus ACGUUAUCCUUGUUGAGAGUUUCGCAAUAUAUUUGUAUUGCAGCAGUCUCAACGAAAGCGUCGAAGCAACCAAAUCGCAUC______GAACA__CCCGACAAAU_____ (((((....(((((((((.(...(((((((....)))))))...).))))))))).)))))................................................ (-18.45 = -19.12 + 0.67)

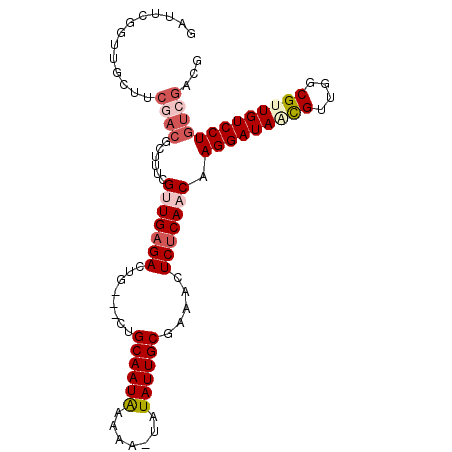
| Location | 4,571,687 – 4,571,782 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -23.23 |
| Energy contribution | -24.45 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
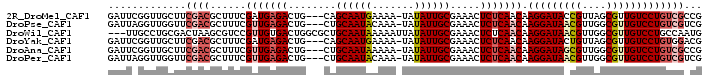
>2R_DroMel_CAF1 4571687 95 + 20766785 GAUUCGGUUGCUUCGACGCUUUCGAUGAGACUG---CAGCAAUGAAAA-UAUAUUGCGAAACUCUCAACAAGGAUACCGUUAGCGUUGUCCUGUCGCCG ....((((.((..(((((((..((.(((((...---..((((((....-..)))))).....))))).)..((...)))..)))))))....)).)))) ( -27.60) >DroPse_CAF1 8432 95 + 1 GAUUAGGUUGGUUCGACGCUUUCGUUGAGACUG---CUGCAAUACAAA-UAUAUUGCGAAACUCUCAACAAGGAUAACGUUGGCGUUGUCCUGUCGUCG .............((((((....(((((((.(.---.(((((((....-..)))))))..).))))))).(((((((((....)))))))))).))))) ( -34.20) >DroWil_CAF1 10930 96 + 1 ---UUGCCUGCGACUAAGCGUCCGUUGUGACUGGCGCUGCAAUAAAAAUUAUAUUGCGAAACUCUCAACAAGGAUAACGUUGGCGUUGUCCUGCCAAUG ---......((((...((((((.((....)).))))))((((((.......)))))).......))....(((((((((....)))))))))))..... ( -28.00) >DroYak_CAF1 9731 95 + 1 GAUUCGGUUGCUUCGACGCUUUCGAUGAGACUG---CAGCAAUGAAAA-UAUAUUGCGAAACUCUCAACAAGGAUACUGUUAGCGUUGUCCUGUGGACG ...((((..((......))..))))(((((...---..((((((....-..)))))).....)))))(((.(((((.((....)).))))))))..... ( -23.80) >DroAna_CAF1 10723 95 + 1 GAUUCGGUUGCUUCGACGCUUUCGUUGAGACUG---CUGCAAUAAAAA-UAUAUUGCGAAACUCUCAACAAGGAUAGCGUUGGCGUUGUCCUGUCGCCG ....((((.((............(((((((.(.---.(((((((....-..)))))))..).))))))).(((((((((....))))))))))).)))) ( -34.40) >DroPer_CAF1 8545 95 + 1 GAUUAGGUUGGUUCGACGCUUUCGUUGAGACUG---CUGCAAUACAAA-UAUAUUGCGAAACUCUCAACAAGGAUAACGUUGGCGUUGUCCUGUCGUCG .............((((((....(((((((.(.---.(((((((....-..)))))))..).))))))).(((((((((....)))))))))).))))) ( -34.20) >consensus GAUUCGGUUGCUUCGACGCUUUCGUUGAGACUG___CUGCAAUAAAAA_UAUAUUGCGAAACUCUCAACAAGGAUAACGUUGGCGUUGUCCUGUCGACG .............((((......(((((((........((((((.......)))))).....))))))).(((((((((....)))))))))))))... (-23.23 = -24.45 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:38 2006