
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,558,485 – 4,558,745 |
| Length | 260 |
| Max. P | 0.916967 |

| Location | 4,558,485 – 4,558,602 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 72.12 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.37 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4558485 117 - 20766785 CCGUCAGCCCAUCAGUACAUUCAACAAUCCAC-CCA-UUUCAGGGUCUCCUGGCGGCCAUUCCCCUGGCCUACAUCCUGCCCGGCCUGGCCUACAUUCAGAUGGAGCCGCACGCCCUGC .((((.(((((((.................((-((.-.....))))....((..(((((....((.(((.........))).))..)))))..))....))))).)).).)))...... ( -31.70) >DroVir_CAF1 37440 105 - 1 -------------AUUUCAUGUAAAUUUUCUCAACA-ACUUAGGGCCUGCUGGCAGCCAUACCCCUGGCCUAUAUACUGCCCGGCCUGGCCUACAUACAAAUGGAACCGCAUGCCCUCU -------------....(((((..............-.....(((((.(((((((((((......))))........)).)))))..))))).(((....))).....)))))...... ( -25.80) >DroPse_CAF1 32148 90 - 1 ----------------------------GCAC-GCAUUUUUAGGGUCUCCUGGCAGCCAUACCCCUGGCUUAUAUACUGCCCGGGCUGGCCUACAUCCGGAUGGAGCCGCACGCCCUCU ----------------------------....-........(((((....((((.((((...(((.(((.........))).))).)))).....(((....)))))))...))))).. ( -29.00) >DroGri_CAF1 36304 104 - 1 -------------AUUUCAUGCAAAUUUCUUC-ACA-ACUUAGGGUCUGCUGGCAGCCAUACCACUGGCCUAUAUACUGCCCGGCCUGGCCUACAUACAAAUGGAACCGCAUGCGCUCU -------------....(((((..........-...-.....(((((.(((((((((((......))))........)).)))))..))))).(((....))).....)))))...... ( -25.50) >DroYak_CAF1 32228 109 - 1 CCAUCCGCC--------CAUUCACCAGUCCAC-CCA-UUUCAGGGUCUCCUGGCGGCCAUACCCCUGGCCUACAUCCUGCCCGGCCUGGCCUACAUCCAGAUGGAGCCGCACGCCCUGC ......(((--------((((..((((...((-((.-.....))))...)))).(((((....((.(((.........))).))..)))))........))))).)).(((.....))) ( -32.00) >DroAna_CAF1 32333 91 - 1 --------------------------GUUGGA-ACA-UUUCAGGGUCUCCUGGCCGCCAUCCCACUGGCCUACAUCCUACCCGGCCUGGCCUACAUCCAGAUGGAGCCGCACGCCCUCC --------------------------......-...-....(((((....((((..(((((.....((((.....((.....))...))))........))))).))))...))))).. ( -25.32) >consensus _________________CAU_CAA__GUCCAC_ACA_UUUCAGGGUCUCCUGGCAGCCAUACCCCUGGCCUACAUACUGCCCGGCCUGGCCUACAUCCAGAUGGAGCCGCACGCCCUCC .........................................(((((....((((.((((..((...(((.........))).))..))))...(((....)))..))))...))))).. (-18.84 = -19.37 + 0.53)

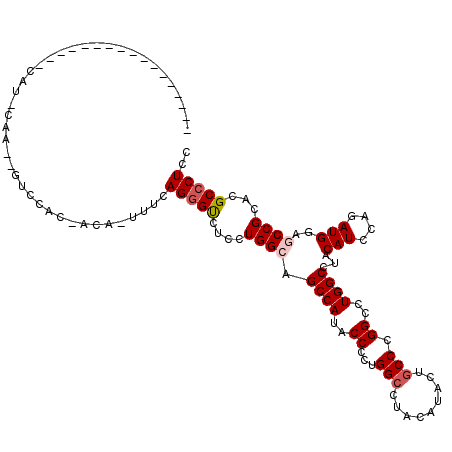
| Location | 4,558,525 – 4,558,641 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -18.72 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4558525 116 - 20766785 UUAUGCCCCACCCAUCAGCCAGCCCAUCAGAACAUCAGCCCGUCAGCCCAUCAGUACAUUCAACAAUCCACCCAUUUCAGGGUCUCCUGGCGGCCAUUCCCCUGGCCUACAUCCUG .................(((((...............((......))......................((((......))))...)))))(((((......)))))......... ( -22.20) >DroSec_CAF1 31845 88 - 1 UUAUGCCCCACCCAUCA----------------------------GCACAUCAGUCCAUUCAACAAUCCACCCAUUUCAGGGUCUCCUGGCGGCCAUUCCCCUGGCCUACAUCCUG ....(((..........----------------------------(.((....)).)............((((......)))).....)))(((((......)))))......... ( -16.80) >DroSim_CAF1 32265 88 - 1 UUAUGCCCCACCCAUCA----------------------------GCACAUCAGUCCAUUCAACAAUCCACCCAUUUCAGGGUCUCCUGGCGGCCAUUCCCCUGGCCUACAUCCUG ....(((..........----------------------------(.((....)).)............((((......)))).....)))(((((......)))))......... ( -16.80) >DroEre_CAF1 32362 96 - 1 UUAUGCCCCACCCAUUAG--------------------CCCAUCAGCCCAUCAGUCCAUUUACGAGUCCACCCAUUUCAGGGUCUUCUGGCGGCCAUACCCCUGGCCUACAUCCUG .................(--------------------((.....((((....((......))(((.........))).)))).....)))(((((......)))))......... ( -18.70) >DroYak_CAF1 32268 88 - 1 UUAUGCCCCACCCAUUAG--------------------CCCAUCCGCC--------CAUUCACCAGUCCACCCAUUUCAGGGUCUCCUGGCGGCCAUACCCCUGGCCUACAUCCUG ..................--------------------..........--------......((((...((((......))))...)))).(((((......)))))......... ( -19.10) >consensus UUAUGCCCCACCCAUCAG____________________CCC_UC_GCCCAUCAGUCCAUUCAACAAUCCACCCAUUUCAGGGUCUCCUGGCGGCCAUUCCCCUGGCCUACAUCCUG .............................................................................(((((((....)))(((((......))))).....)))) (-15.00 = -15.00 + 0.00)


| Location | 4,558,602 – 4,558,705 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.69 |
| Mean single sequence MFE | -16.12 |
| Consensus MFE | -14.93 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4558602 103 - 20766785 CGCCAAUCCCAAACGGCAUCUACAAAUUCUGGCCGGAACCCGUCUUUGUAUUUGCCAUGCAAAUUUAUGCCCCACCCAUCAGCCAGCCCAUCAGAACAUCAGC .((...........((((..((((((..(.((......)).)..))))))..))))..(((......)))...........)).................... ( -16.70) >DroSim_CAF1 32336 81 - 1 CGCCAAUCCCAAACGGCAUCUACAAAUUCUGGCCCGAACCCGGCUUUGUAUUUGCCAUGCAAAUUUAUGCCCCACCCAUCA---------------------- ..............((((..((((((..((((.......)))).))))))..))))..(((......)))...........---------------------- ( -16.60) >DroEre_CAF1 32439 83 - 1 UGUCAAUCCCAAACGGCAUAUACAAAUUCUGGCCUGAACCCGGCUUUGUAUUUGCCAUGCAAAUUUAUGCCCCACCCAUUAG--------------------C ..............((((.(((((((..((((.......)))).))))))).))))..(((......)))............--------------------. ( -17.60) >DroYak_CAF1 32337 82 - 1 UGCCAAUCCCAAACGGCAUCUACAAAUUCUGGCCCGAACCC-GCUUUGUAUUUGCCAUGCAAAUUUAUGCCCCACCCAUUAG--------------------C ..............((((..((((((..(.((......)).-).))))))..))))..(((......)))............--------------------. ( -13.60) >consensus CGCCAAUCCCAAACGGCAUCUACAAAUUCUGGCCCGAACCCGGCUUUGUAUUUGCCAUGCAAAUUUAUGCCCCACCCAUCAG____________________C ..............((((..((((((..(.((......)).)..))))))..))))..(((......)))................................. (-14.93 = -15.17 + 0.25)

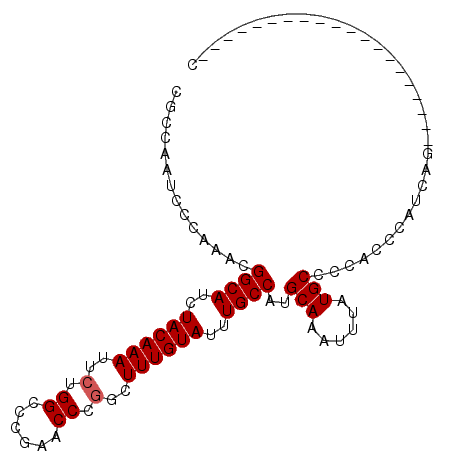

| Location | 4,558,641 – 4,558,745 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -13.45 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4558641 104 - 20766785 CAUUUGCCCCUUGUAUUCCAGUCACGGGUAUUGGCCACAACGCCAAUCCCAAACGGCAUCUACAAAUUCUGGCCGGAACCCGUCUUUG-----------UAUUUGCCAUGCAAAU .((((((..................(((.((((((......)))))))))....((((..((((((..(.((......)).)..))))-----------))..))))..)))))) ( -32.50) >DroSec_CAF1 31933 91 - 1 CAUUUGGCUCUUGUAUUCCGGUCACGGGUAUUGGCCACAA-------------CGGCAUCUACAAAUUCAGGCCCGAACCCGGCUUUG-----------UAUUUGCCAUGCAAAU ....(((((...((((.(((....))))))).)))))...-------------.((((..((((((..(.((......)).)..))))-----------))..))))........ ( -27.30) >DroSim_CAF1 32353 104 - 1 CAUUUGGCUCUUGUAUUCCGGUCACGGGUAUUGGCCACAACGCCAAUCCCAAACGGCAUCUACAAAUUCUGGCCCGAACCCGGCUUUG-----------UAUUUGCCAUGCAAAU ..........((((((.........(((.((((((......)))))))))....((((..((((((..((((.......)))).))))-----------))..)))))))))).. ( -32.20) >DroEre_CAF1 32458 104 - 1 CAUUUGUCCCUUGUAUUCUGGUCACGGGUAUUGGCCACAAUGUCAAUCCCAAACGGCAUAUACAAAUUCUGGCCUGAACCCGGCUUUG-----------UAUUUGCCAUGCAAAU ..........((((((.........(((.((((((......)))))))))....((((.(((((((..((((.......)))).))))-----------))).)))))))))).. ( -30.40) >DroYak_CAF1 32356 103 - 1 CAUUCGGCCCUUGUAUUCCGGUCACGGGUAUUGGCCACAAUGCCAAUCCCAAACGGCAUCUACAAAUUCUGGCCCGAACCC-GCUUUG-----------UAUUUGCCAUGCAAAU .....((((..........))))..(((.((((((......)))))))))....((((..((((((..(.((......)).-).))))-----------))..))))........ ( -29.50) >DroAna_CAF1 32434 108 - 1 UUUUUCCCCCUUGCAUUCUGGUCACGGGUAUCGGCCAAAAUGGCCUCCC-------CCUCCACACAUUUUCCCCCAUUUCGGCCUUUGUAGUUUGUGGGUAUUUGGCAUGCAAAU ..........((((((.(..(..(((((....((((.....))))...)-------)).(((((.(((.....((.....)).......))).)))))))..)..).)))))).. ( -28.50) >consensus CAUUUGGCCCUUGUAUUCCGGUCACGGGUAUUGGCCACAAUGCCAAUCCCAAACGGCAUCUACAAAUUCUGGCCCGAACCCGGCUUUG___________UAUUUGCCAUGCAAAU ..........((((((.........((((...(((((................................)))))...))))(((....................))))))))).. (-13.45 = -14.12 + 0.67)

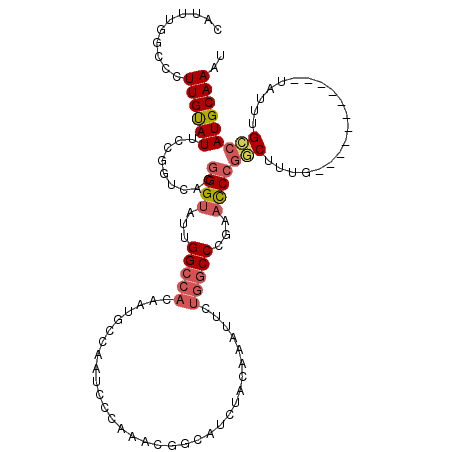
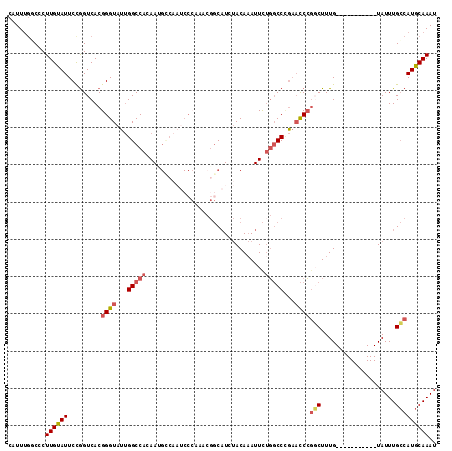
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:33 2006