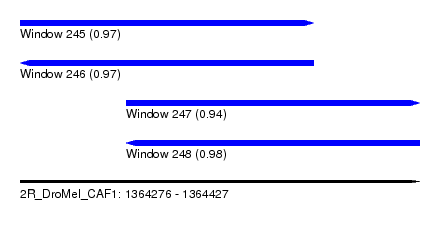
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,364,276 – 1,364,427 |
| Length | 151 |
| Max. P | 0.978627 |

| Location | 1,364,276 – 1,364,387 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 93.95 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.84 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1364276 111 + 20766785 GAAUAGAAUCCGUUCGUUUUGUUUCUUUUUUGCGUGCCAAAUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGCAACGAUUUUUCGGAACUAG ((((.......))))((((((.....((.((((((((.((((..((....))..))))))))......((((((((......)))))))))))).)).....))))))... ( -28.40) >DroSec_CAF1 245 111 + 1 GACUAGAAUCCGUUCGUUUUGUUUCUUUUUUGCGUGCCAAAUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGCAACGAUUUUUCGGAACUAG ..((((..((((.((((.............(((((((.((((..((....))..))))))))......((((((((......))))))))))))))).....)))).)))) ( -29.51) >DroEre_CAF1 907 106 + 1 -----GAAUCCGUUCGUUUUGUUUCAUUUUUGCGUGCCAAAUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGCAACGAUUUUUCGGAACUAG -----......((((..............((((((((.((((..((....))..))))))))......((((((((......))))))))))))(((....)))))))... ( -28.40) >DroYak_CAF1 544 106 + 1 -----CAAUCCGUUCGUUUUGUUUCAUUUUUGCUUGCCAAAUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGGAAGGUUUUUUCGGAACUAG -----...............(((((......((((.((..((((((((.((.......))))))))))((((((((......)))))))))).))))......)))))... ( -26.50) >consensus _____GAAUCCGUUCGUUUUGUUUCAUUUUUGCGUGCCAAAUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGCAACGAUUUUUCGGAACUAG ...............((((((........((((((((.((((..((....))..))))))))......((((((((......))))))))))))........))))))... (-24.34 = -24.84 + 0.50)

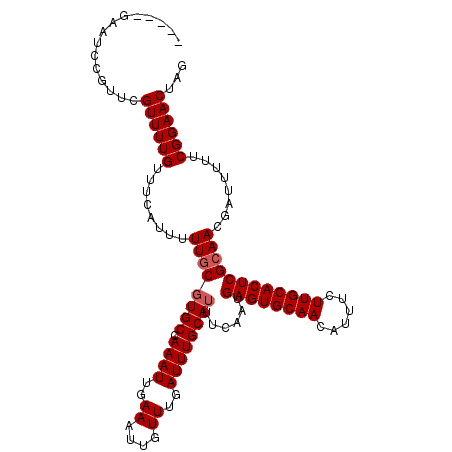
| Location | 1,364,276 – 1,364,387 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 93.95 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1364276 111 - 20766785 CUAGUUCCGAAAAAUCGUUGCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAUUUGGCACGCAAAAAAGAAACAAAACGAACGGAUUCUAUUC .(((.((((.....(((((((((((((((......))))))))((((((((............))))))))...)).(........)......))))).))))..)))... ( -25.20) >DroSec_CAF1 245 111 - 1 CUAGUUCCGAAAAAUCGUUGCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAUUUGGCACGCAAAAAAGAAACAAAACGAACGGAUUCUAGUC ((((.((((.....(((((((((((((((......))))))))((((((((............))))))))...)).(........)......))))).))))..)))).. ( -27.70) >DroEre_CAF1 907 106 - 1 CUAGUUCCGAAAAAUCGUUGCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAUUUGGCACGCAAAAAUGAAACAAAACGAACGGAUUC----- ...(((((((....)))((((((((((((......))))))))((((((((............)))))))).......))))..............))))......----- ( -24.60) >DroYak_CAF1 544 106 - 1 CUAGUUCCGAAAAAACCUUCCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAUUUGGCAAGCAAAAAUGAAACAAAACGAACGGAUUG----- .....((((.......(((((((((((((......))))))))((((((((............))))))))..)).)))((....))............))))...----- ( -22.20) >consensus CUAGUUCCGAAAAAUCGUUGCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAUUUGGCACGCAAAAAAGAAACAAAACGAACGGAUUC_____ .....((((........((((((((((((......))))))))((((((((............)))))))).......))))........(.....)..))))........ (-21.52 = -21.77 + 0.25)

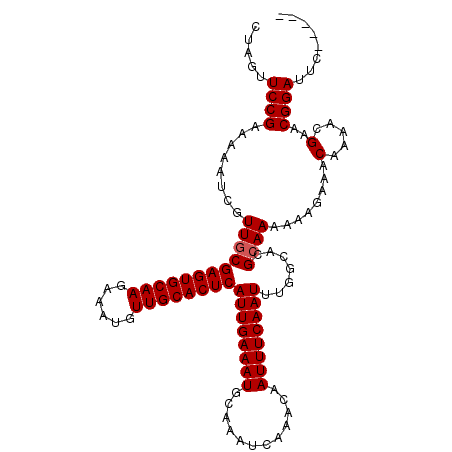
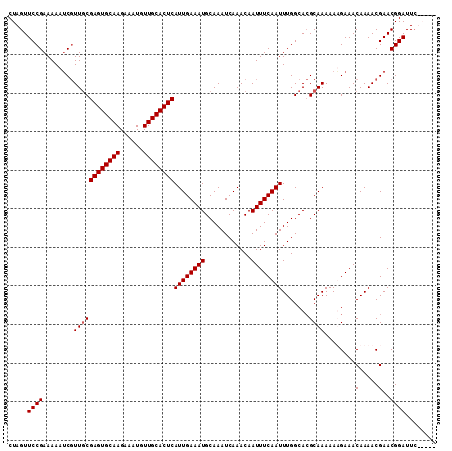
| Location | 1,364,316 – 1,364,427 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -30.90 |
| Energy contribution | -31.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1364316 111 + 20766785 AUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGCAACGAUUUUUCGGAACUAGCUGGGAGGAAAGUAAUUCCUGCUCGCAGUUUGCUGAAGCA ((((((((.((.......))))))))))((((((((......))))))))((..(((....)))(((((.((.((.(((((.....))))).)).)))))))......)). ( -36.00) >DroSec_CAF1 285 111 + 1 AUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGCAACGAUUUUUCGGAACUAGCUGGGAGGAAAGUAAUUCCUGCUCGCAGUUUGCUGAAGCA ((((((((.((.......))))))))))((((((((......))))))))((..(((....)))(((((.((.((.(((((.....))))).)).)))))))......)). ( -36.00) >DroEre_CAF1 942 111 + 1 AUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGCAACGAUUUUUCGGAACUAGCUGGGAAGAAAGUAAUUCUUGCUCGCAGUUUGCUGAAGCA ((((((((.((.......))))))))))((((((((......))))))))((..(((....)))(((((.((.((.(((((.....))))).)).)))))))......)). ( -33.60) >DroYak_CAF1 579 111 + 1 AUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGGAAGGUUUUUUCGGAACUAGCUGGGAAAAAAGUAAUUCCUGCUCGCAGUUUGCUGAAGCA ((((((((.((.......))))))))))((((((((......))))))))((((..((((..(((......)))..))))......)))).(((..(((....))).))). ( -32.80) >consensus AUUGAAAUUGUUUGAUUUGCAUUUCAAUGAGUGCAACAUUUCUUGCACUCGCAACGAUUUUUCGGAACUAGCUGGGAAGAAAGUAAUUCCUGCUCGCAGUUUGCUGAAGCA ((((((((.((.......))))))))))((((((((......)))))))).........((((((.(((.((.((.(((((.....))))).)).)))))...)))))).. (-30.90 = -31.15 + 0.25)

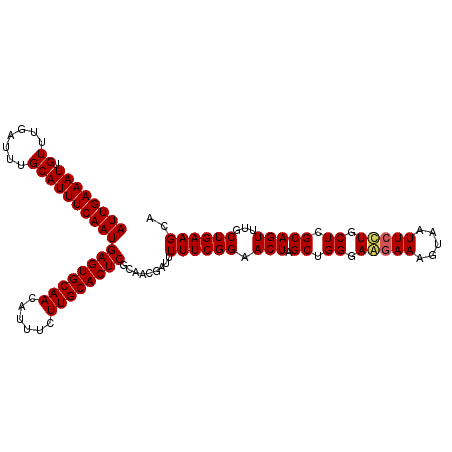
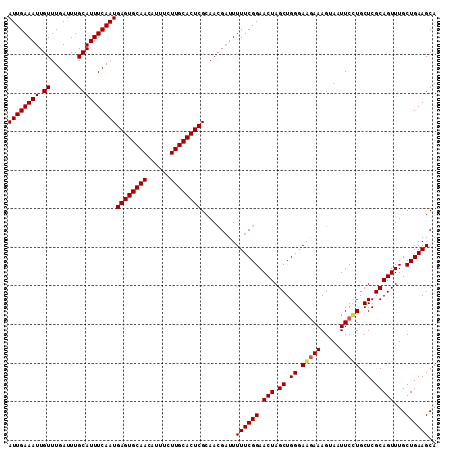
| Location | 1,364,316 – 1,364,427 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -27.62 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1364316 111 - 20766785 UGCUUCAGCAAACUGCGAGCAGGAAUUACUUUCCUCCCAGCUAGUUCCGAAAAAUCGUUGCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAU ((((((((....))).)))))((((.....)))).........((..(((....)))..))((((((((......))))))))((((((((............)))))))) ( -29.90) >DroSec_CAF1 285 111 - 1 UGCUUCAGCAAACUGCGAGCAGGAAUUACUUUCCUCCCAGCUAGUUCCGAAAAAUCGUUGCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAU ((((((((....))).)))))((((.....)))).........((..(((....)))..))((((((((......))))))))((((((((............)))))))) ( -29.90) >DroEre_CAF1 942 111 - 1 UGCUUCAGCAAACUGCGAGCAAGAAUUACUUUCUUCCCAGCUAGUUCCGAAAAAUCGUUGCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAU .(((((((....))).))))(((((.....)))))........((..(((....)))..))((((((((......))))))))((((((((............)))))))) ( -27.40) >DroYak_CAF1 579 111 - 1 UGCUUCAGCAAACUGCGAGCAGGAAUUACUUUUUUCCCAGCUAGUUCCGAAAAAACCUUCCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAU ((((((((....))).)))))(((((((((........)).))))))).............((((((((......))))))))((((((((............)))))))) ( -29.30) >consensus UGCUUCAGCAAACUGCGAGCAGGAAUUACUUUCCUCCCAGCUAGUUCCGAAAAAUCGUUGCGAGUGCAAGAAAUGUUGCACUCAUUGAAAUGCAAAUCAAACAAUUUCAAU ((((((((....))).)))))(((((((((........)).))))))).............((((((((......))))))))((((((((............)))))))) (-27.62 = -27.88 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:46 2006