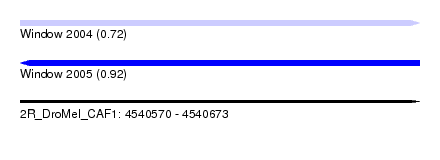
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,540,570 – 4,540,673 |
| Length | 103 |
| Max. P | 0.923428 |

| Location | 4,540,570 – 4,540,673 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.95 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
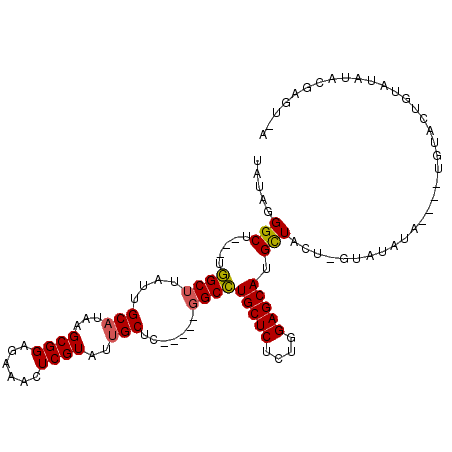
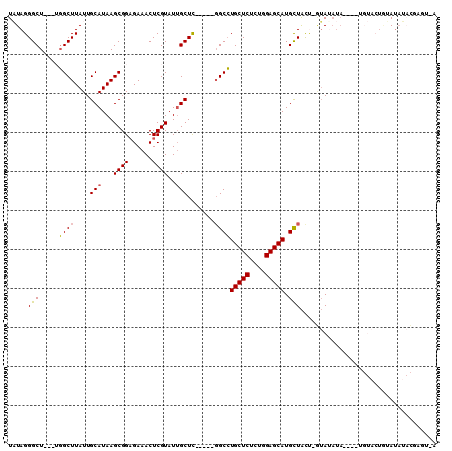
>2R_DroMel_CAF1 4540570 103 + 20766785 U-ACUCGUAUUUAUA-UACAAAUGUAUGUACAAGUAGCAUGCUCCAGAGAGCAGGCC-----GAGCAAUACGAGUUUCUCCGCUUAUGCAAUAAGCCAGCCAGCCCUAUA .-(((((((((((((-(((....)))))))......((.(((((....))))).)).-----....)))))))))......((((((...)))))).............. ( -26.80) >DroSec_CAF1 14176 99 + 1 U-ACUCGUAUAUACAGUACA----UAUAUAC-AGUAGCAUGCUCCAGAGAGCAGGCC-----GAGCAAUACGAGUUUCGCCGCUUAUGCAAUAAGCCAGCCAGCCCUAUA (-(((.(((((((.......----)))))))-))))((.(((((....)))))(((.-----((((.......)))).)))((((((...))))))..)).......... ( -25.20) >DroSim_CAF1 14190 100 + 1 U-ACUCGUAUAUACAGUACA----UAUAUAC-AGUAGCAUGCUCCAGAGAGCAGGCC----CAAGCAAUACGAGUUUCUCCGCUUAUGCAAUAAGCCAGCCAGCCCUAUA .-((((((((.(((.(((..----....)))-.)))((.(((((....))))).)).----......))))))))......((((((...)))))).............. ( -23.00) >DroYak_CAF1 14057 97 + 1 U-UCGAGUAUAU---GUACG----UAUAUAC-AGUAGCAUGCUCCAGAGAGCAGGCCGAGCAGAGCAAUACGAGUUUCUCCGCUUAUGCAAUAAGCC----AGCCCUAUA .-....((((((---(....----)))))))-.((((..(((((....)))))(((.((((.(((.(((....))).))).))))..((.....)).----.))))))). ( -23.80) >DroMoj_CAF1 17547 88 + 1 CAACGCAGCAAUUCA------------AUAU--AUAACAUGCUCUAAAGAGCAUGCA-----GAGCAAUACGAGUUUCUCCGCUUAUGCAAUAAGCAA---AGCCCUAAA ....(.((.(((((.------------....--....(((((((....)))))))..-----.(....)..))))).)).)((((.(((.....))))---)))...... ( -18.40) >DroAna_CAF1 15284 82 + 1 --------AUAU---G--------UAUUUCAAGACAACAUGCUCCAAAGAGCAGGCC-----AAGCCAUACGAGUUUCUCCGCUUAUGCAAUAAGCC----AGCCCUAAA --------...(---(--------(((............(((((....)))))(((.-----..)))))))).........((((((...)))))).----......... ( -14.50) >consensus U_ACUCGUAUAUACAGUACA____UAUAUAC_AGUAGCAUGCUCCAGAGAGCAGGCC_____GAGCAAUACGAGUUUCUCCGCUUAUGCAAUAAGCCA___AGCCCUAUA .......................................(((((....)))))(((......((((.......))))....((((((...))))))......)))..... (-12.92 = -12.87 + -0.06)

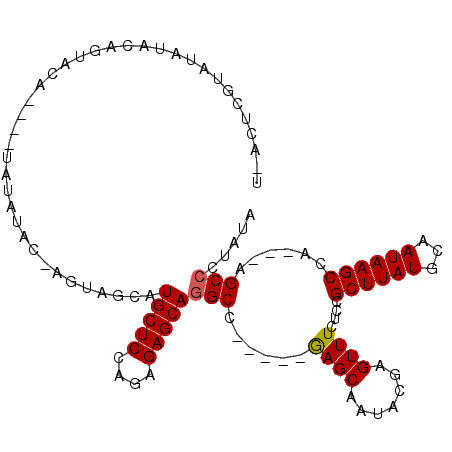
| Location | 4,540,570 – 4,540,673 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.95 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.48 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
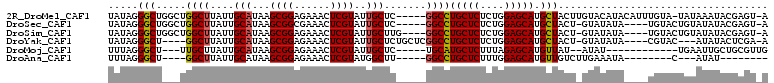
>2R_DroMel_CAF1 4540570 103 - 20766785 UAUAGGGCUGGCUGGCUUAUUGCAUAAGCGGAGAAACUCGUAUUGCUC-----GGCCUGCUCUCUGGAGCAUGCUACUUGUACAUACAUUUGUA-UAUAAAUACGAGU-A ..........(((.((.....))...)))......(((((((((....-----(((.(((((....))))).)))...((((((......))))-))..)))))))))-. ( -31.90) >DroSec_CAF1 14176 99 - 1 UAUAGGGCUGGCUGGCUUAUUGCAUAAGCGGCGAAACUCGUAUUGCUC-----GGCCUGCUCUCUGGAGCAUGCUACU-GUAUAUA----UGUACUGUAUAUACGAGU-A ((..((..(.((((.(((((...))))))))).)..))..)).(((((-----(((.(((((....))))).)))...-(((((((----(.....))))))))))))-) ( -33.00) >DroSim_CAF1 14190 100 - 1 UAUAGGGCUGGCUGGCUUAUUGCAUAAGCGGAGAAACUCGUAUUGCUUG----GGCCUGCUCUCUGGAGCAUGCUACU-GUAUAUA----UGUACUGUAUAUACGAGU-A .....((.((((.((((((..(((...((((......))))..))).))----))))(((((....))))).))))))-(((((((----(.....))))))))....-. ( -30.60) >DroYak_CAF1 14057 97 - 1 UAUAGGGCU----GGCUUAUUGCAUAAGCGGAGAAACUCGUAUUGCUCUGCUCGGCCUGCUCUCUGGAGCAUGCUACU-GUAUAUA----CGUAC---AUAUACUCGA-A ....(((((----(((.....))...(((((((............)))))))))))))((((....))))........-((((((.----.....---))))))....-. ( -28.00) >DroMoj_CAF1 17547 88 - 1 UUUAGGGCU---UUGCUUAUUGCAUAAGCGGAGAAACUCGUAUUGCUC-----UGCAUGCUCUUUAGAGCAUGUUAU--AUAU------------UGAAUUGCUGCGUUG ...(((((.---.(((....(((....)))(((...))))))..))))-----)((((((((....))))))))...--....------------............... ( -23.10) >DroAna_CAF1 15284 82 - 1 UUUAGGGCU----GGCUUAUUGCAUAAGCGGAGAAACUCGUAUGGCUU-----GGCCUGCUCUUUGGAGCAUGUUGUCUUGAAAUA--------C---AUAU-------- ((((((((.----((((....((....((((......))))...))..-----))))(((((....)))))....))))))))...--------.---....-------- ( -23.50) >consensus UAUAGGGCU___UGGCUUAUUGCAUAAGCGGAGAAACUCGUAUUGCUC_____GGCCUGCUCUCUGGAGCAUGCUACU_GUAUAUA____UGUACUGUAUAUACGAGU_A .....(((.....((((....(((...((((......))))..))).......))))(((((....))))).)))................................... (-17.65 = -17.48 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:27 2006