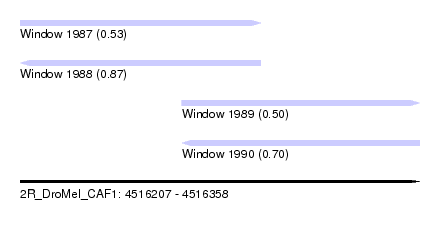
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,516,207 – 4,516,358 |
| Length | 151 |
| Max. P | 0.869051 |

| Location | 4,516,207 – 4,516,298 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -29.67 |
| Energy contribution | -29.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
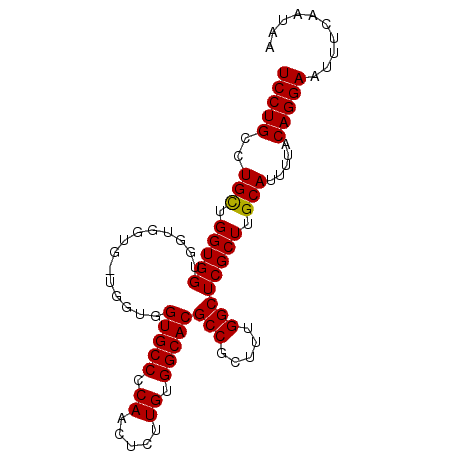
>2R_DroMel_CAF1 4516207 91 + 20766785 AAAAGCAGAAGCAGUCGGCAGUGGCAGCGCAAUUGAAACACUUGUACUAAGCUGCUGUUGCUCCUGC-UGUUGGUGGUGGUGGUGGUGCUGG ...(((((.(((((.(((((((...((..(((.((...)).)))..))..)))))))))))).))))-).(..(((.(......).)))..) ( -29.80) >DroSec_CAF1 50592 86 + 1 AAAAGCAGAAGCAGUCGGCAGUGGCAGCGCAAUUGAAACACUUGUACUAAGCUGCUGCUGCUCCUGCCUGCUGGUGGUGGUG------GUGG ...(((((..((((..(((((..(((((((((.((...)).)))).....)))))..))))).)))))))))..........------.... ( -33.10) >DroSim_CAF1 49509 92 + 1 AAAAGCAGAAGCAGUCGGCAGUGGCAGCGCAAUUGAAACACUUGUACUAAGCUGCUGUUGCUCCUGCCUGCUGGUGGUGGUGGUGCUGGUGG ....((((.(((((.(((((((...((..(((.((...)).)))..))..)))))))))))).))))(..(..((.........))..)..) ( -32.80) >consensus AAAAGCAGAAGCAGUCGGCAGUGGCAGCGCAAUUGAAACACUUGUACUAAGCUGCUGUUGCUCCUGCCUGCUGGUGGUGGUGGUG_UGGUGG ...(((((..((((..(((((..(((((((((.((...)).)))).....)))))..))))).))))))))).................... (-29.67 = -29.57 + -0.11)



| Location | 4,516,207 – 4,516,298 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -23.27 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4516207 91 - 20766785 CCAGCACCACCACCACCACCAACA-GCAGGAGCAACAGCAGCUUAGUACAAGUGUUUCAAUUGCGCUGCCACUGCCGACUGCUUCUGCUUUU .......................(-(((((((((.(.((((..(((..(((.........)))..)))...)))).)..))))))))))... ( -23.90) >DroSec_CAF1 50592 86 - 1 CCAC------CACCACCACCAGCAGGCAGGAGCAGCAGCAGCUUAGUACAAGUGUUUCAAUUGCGCUGCCACUGCCGACUGCUUCUGCUUUU ....------.............(((((((((((((.((((..(((..(((.........)))..)))...)))).).)))))))))))).. ( -29.20) >DroSim_CAF1 49509 92 - 1 CCACCAGCACCACCACCACCAGCAGGCAGGAGCAACAGCAGCUUAGUACAAGUGUUUCAAUUGCGCUGCCACUGCCGACUGCUUCUGCUUUU ......((.............))(((((((((((.(.((((..(((..(((.........)))..)))...)))).)..))))))))))).. ( -24.52) >consensus CCACCA_CACCACCACCACCAGCAGGCAGGAGCAACAGCAGCUUAGUACAAGUGUUUCAAUUGCGCUGCCACUGCCGACUGCUUCUGCUUUU .......................(((((((((((...((((..(((..(((.........)))..)))...))))....))))))))))).. (-23.27 = -23.60 + 0.33)



| Location | 4,516,268 – 4,516,358 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -25.99 |
| Energy contribution | -25.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4516268 90 + 20766785 UCCUGC-UGUUGGUGGUGGUGGUGGUGCUGGUGCCCCAACUCUUGUGGCACGCCGCUUUGGCUCGCUUGCAUUUUACAGGAAUUUCAAUAA (((((.-(((.((((((.(.((((((....(((((.((.....)).))))))))))).).)).)))).))).....))))).......... ( -30.10) >DroSec_CAF1 50653 85 + 1 UCCUGCCUGCUGGUGGUGGUG------GUGGUGCCACAACUCUUGUGGCACGCCGCUUUGGCUCGCUUGCAUUUUACAGGAAUUUCAAUAA (((((..(((.((((((((((------(((.(((((((.....))))))))))))))...)).)))).))).....))))).......... ( -35.50) >DroSim_CAF1 49570 91 + 1 UCCUGCCUGCUGGUGGUGGUGGUGCUGGUGGUGCCCCAACUCUUGUGGCACGCCGCUUUGGCUCGCUUGCAUUUUACAGGAAUUUCAAUAA (((((((....))).((((.(((((.(((((((((.((.....)).)))))(((.....))).)))).))))))))))))).......... ( -30.50) >consensus UCCUGCCUGCUGGUGGUGGUGGUG_UGGUGGUGCCCCAACUCUUGUGGCACGCCGCUUUGGCUCGCUUGCAUUUUACAGGAAUUUCAAUAA (((((..(((.(((((..............(((((.((.....)).)))))(((.....)))))))).))).....))))).......... (-25.99 = -25.77 + -0.22)

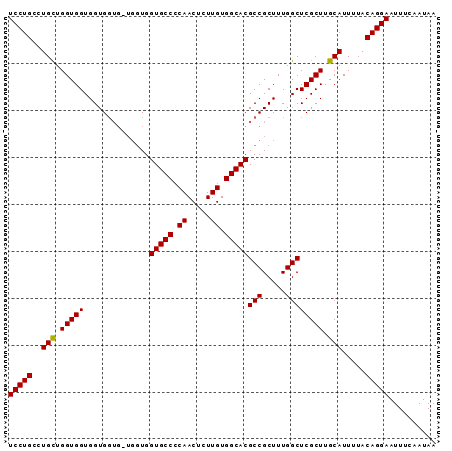
| Location | 4,516,268 – 4,516,358 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -22.87 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
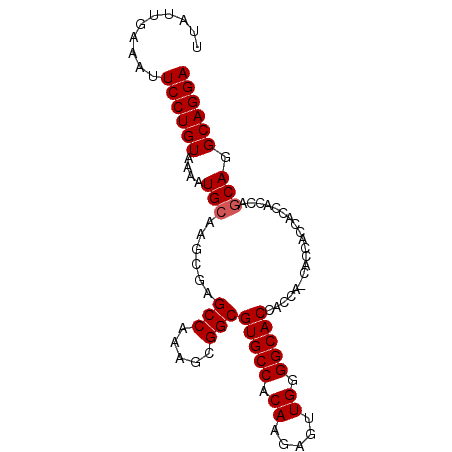
>2R_DroMel_CAF1 4516268 90 - 20766785 UUAUUGAAAUUCCUGUAAAAUGCAAGCGAGCCAAAGCGGCGUGCCACAAGAGUUGGGGCACCAGCACCACCACCACCACCAACA-GCAGGA ..........((((((.........(((.(((.....))).))).......(((((((.................)).))))).-)))))) ( -22.53) >DroSec_CAF1 50653 85 - 1 UUAUUGAAAUUCCUGUAAAAUGCAAGCGAGCCAAAGCGGCGUGCCACAAGAGUUGUGGCACCAC------CACCACCACCAGCAGGCAGGA ..........((((((....(((......(((.....)))((((((((.....))))))))...------...........))).)))))) ( -30.10) >DroSim_CAF1 49570 91 - 1 UUAUUGAAAUUCCUGUAAAAUGCAAGCGAGCCAAAGCGGCGUGCCACAAGAGUUGGGGCACCACCAGCACCACCACCACCAGCAGGCAGGA ..........((((((....(((..(((.(((.....))).))).......((((((....).))))).............))).)))))) ( -26.90) >consensus UUAUUGAAAUUCCUGUAAAAUGCAAGCGAGCCAAAGCGGCGUGCCACAAGAGUUGGGGCACCACCA_CACCACCACCACCAGCAGGCAGGA ..........((((((....(((......(((.....)))(((((.((.....)).)))))....................))).)))))) (-22.87 = -23.20 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:12 2006