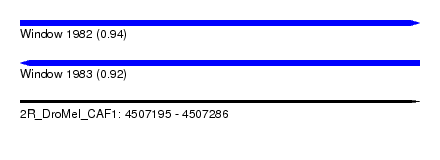
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,507,195 – 4,507,286 |
| Length | 91 |
| Max. P | 0.939424 |

| Location | 4,507,195 – 4,507,286 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -16.25 |
| Energy contribution | -16.62 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
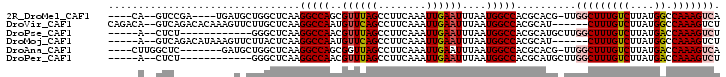
>2R_DroMel_CAF1 4507195 91 + 20766785 ----CA--GUCCGA----UGAUGCUGGCUCAAGGCCAGCGUUUAGCCUUCAAAUUGAAUUUAAUGGCCACGCACG-UUGGCUUUGUCUUAUGGCCAAAGUCA ----..--......----((((..(((((((((((((((((...(((.................))).....)))-)))))))))......)))))..)))) ( -29.93) >DroVir_CAF1 51534 94 + 1 CAGACA--GUCAGACACAAAGUUCUUGCUCAAGGCCAAUGUUCAGCCUUCAAAUUGAAUUUAAUGGCCACGCAU------CUUUGUCUUAUGGCCAAAGUCU .((((.--((((...((((((....(((....(((((..((((((........))))))....)))))..))).------))))))....))))....)))) ( -24.70) >DroPse_CAF1 54206 83 + 1 -----A--CUCU------------GGGCUCAAGGCCAACGUUUAGCCUUCAAAUUGAAUUUAAUGGCCACGCAUGCUUGGCUUUGUCUUAUGACCAAAGUCU -----.--....------------((((....(((((..((((((........))))))....)))))......))))(((((((((....)).))))))). ( -19.40) >DroMoj_CAF1 53606 89 + 1 -----A--GUCAGACAUAAAGUUCUUACUCAAGGCCAAUGUUCAGCCUUCAAAUUGAAUUUAAUGGCCACGCAU------CUUUGUCUUAUGGCCAAAGUCU -----.--...((((...((((((......(((((.........)))))......))))))..((((((.(((.------...)))....))))))..)))) ( -22.30) >DroAna_CAF1 40428 90 + 1 ----CUUGGCUC-------GAUGCUGGCUCAAGGCCAGCGGUUAGCCUUCAAAUUGAAUUUAAUGGCCACGCACG-UUGGCUUUGUCUUAUGACCAAAGUCA ----...((((.-------..(((((((.....)))))))...))))..............((((((...)).))-))(((((((((....)).))))))). ( -29.80) >DroPer_CAF1 57317 83 + 1 -----A--CUCU------------GGGCUCAAGGCCAACGUUUAGCCUUCAAAUUGAAUUUAAUGGCCACGCAUGCUUGGCUUUGUCUUAUGACCAAAGUCU -----.--....------------((((....(((((..((((((........))))))....)))))......))))(((((((((....)).))))))). ( -19.40) >consensus _____A__GUCA_________U_CUGGCUCAAGGCCAACGUUUAGCCUUCAAAUUGAAUUUAAUGGCCACGCAUG_UUGGCUUUGUCUUAUGACCAAAGUCU ................................(((((..((((((........))))))....)))))..........(((((((((....)).))))))). (-16.25 = -16.62 + 0.36)


| Location | 4,507,195 – 4,507,286 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -17.78 |
| Energy contribution | -16.64 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
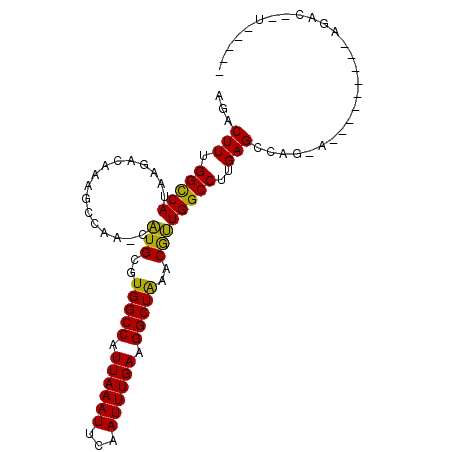
>2R_DroMel_CAF1 4507195 91 - 20766785 UGACUUUGGCCAUAAGACAAAGCCAA-CGUGCGUGGCCAUUAAAUUCAAUUUGAAGGCUAAACGCUGGCCUUGAGCCAGCAUCA----UCGGAC--UG---- .....(((((...........)))))-.((.(((((((.((((((...)))))).)))))...((((((.....))))))....----.)).))--..---- ( -27.40) >DroVir_CAF1 51534 94 - 1 AGACUUUGGCCAUAAGACAAAG------AUGCGUGGCCAUUAAAUUCAAUUUGAAGGCUGAACAUUGGCCUUGAGCAAGAACUUUGUGUCUGAC--UGUCUG ((((..(((((((.........------....))))))).(((((((...((.(((((..(...)..))))).))...))).)))).))))...--...... ( -21.92) >DroPse_CAF1 54206 83 - 1 AGACUUUGGUCAUAAGACAAAGCCAAGCAUGCGUGGCCAUUAAAUUCAAUUUGAAGGCUAAACGUUGGCCUUGAGCCC------------AGAG--U----- ..(((((((.(.((((.....((((...(((..(((((.((((((...)))))).)))))..))))))))))).).))------------))))--)----- ( -24.50) >DroMoj_CAF1 53606 89 - 1 AGACUUUGGCCAUAAGACAAAG------AUGCGUGGCCAUUAAAUUCAAUUUGAAGGCUGAACAUUGGCCUUGAGUAAGAACUUUAUGUCUGAC--U----- ((((..(((((((.........------....))))))).(((((((...((.(((((..(...)..))))).))...))).)))).))))...--.----- ( -22.92) >DroAna_CAF1 40428 90 - 1 UGACUUUGGUCAUAAGACAAAGCCAA-CGUGCGUGGCCAUUAAAUUCAAUUUGAAGGCUAACCGCUGGCCUUGAGCCAGCAUC-------GAGCCAAG---- (((.(((((((....)))...((((.-(....)))))...)))).))).......((((....((((((.....))))))...-------.))))...---- ( -27.10) >DroPer_CAF1 57317 83 - 1 AGACUUUGGUCAUAAGACAAAGCCAAGCAUGCGUGGCCAUUAAAUUCAAUUUGAAGGCUAAACGUUGGCCUUGAGCCC------------AGAG--U----- ..(((((((.(.((((.....((((...(((..(((((.((((((...)))))).)))))..))))))))))).).))------------))))--)----- ( -24.50) >consensus AGACUUUGGCCAUAAGACAAAGCCAA_CAUGCGUGGCCAUUAAAUUCAAUUUGAAGGCUAAACGUUGGCCUUGAGCCAG_A_________AGAC__U_____ ...(((.(((((................(((..(((((.((((((...)))))).)))))..))))))))..)))........................... (-17.78 = -16.64 + -1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:06 2006