
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,506,484 – 4,506,583 |
| Length | 99 |
| Max. P | 0.824739 |

| Location | 4,506,484 – 4,506,583 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -27.35 |
| Energy contribution | -28.85 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
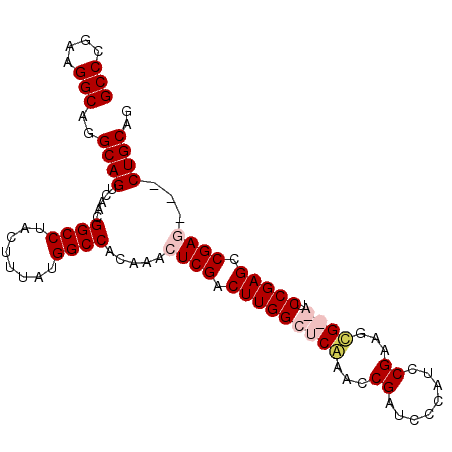
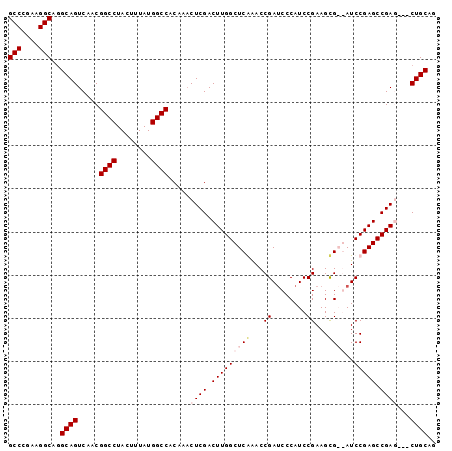
>2R_DroMel_CAF1 4506484 99 + 20766785 GCCCGAAGGCAGGCAGUCAACGGCCUACUUUAUGGCCACAAACUCGACUUGGCUCGAACCGAUCCCAUCCGAAGCGAGAUCCGAGCCGAGCCGCUGCAG (((....)))..(((((....((((........))))........(.(((((((((....((((.(.........).))))))))))))).)))))).. ( -36.40) >DroSec_CAF1 40237 96 + 1 GCCCGAAGGCAGGCAGUCAACGGCCUACUUUAUGGCCACAAACUCGACUUGGCUCGAACCGAUCCCAUCCGAAGCGAGAUCCGAGCCGAA---CUGCAG (((....)))..(((((....((((........))))......(((.(((((((((...((........))...))))..))))).))))---)))).. ( -33.40) >DroEre_CAF1 40175 94 + 1 GCCCGAAGGCAGGCAGUCAACGGCCUACUUUAUGGCCACAAACUCGACUUGGCUCAGACCGAGCCCAUCCGAAGCG--AUCCGAGCCGAG---CUGCAG (((....)))..((((.....((((........))))..........((((((((.((.((..(......)...))--.)).))))))))---)))).. ( -33.30) >DroYak_CAF1 42961 94 + 1 GCCUGAAGGCAAGCAGUCAACGGCCUACUUUAUGGCCCCCACCUCGACUUGGCUCAAACCGAUCCCAUCCGAAGUG--AUCCGAGCCGAC---CUGCAG (((....)))..((((.....((((........))))......(((.(((((.(((...((........))...))--).))))).))).---)))).. ( -30.50) >consensus GCCCGAAGGCAGGCAGUCAACGGCCUACUUUAUGGCCACAAACUCGACUUGGCUCAAACCGAUCCCAUCCGAAGCG__AUCCGAGCCGAG___CUGCAG (((....)))..((((.....((((........)))).....((((.(((((((((...((........))...))))..))))).))))...)))).. (-27.35 = -28.85 + 1.50)

| Location | 4,506,484 – 4,506,583 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -34.19 |
| Energy contribution | -35.25 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
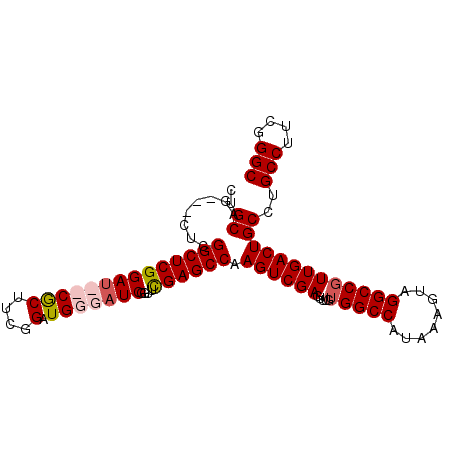
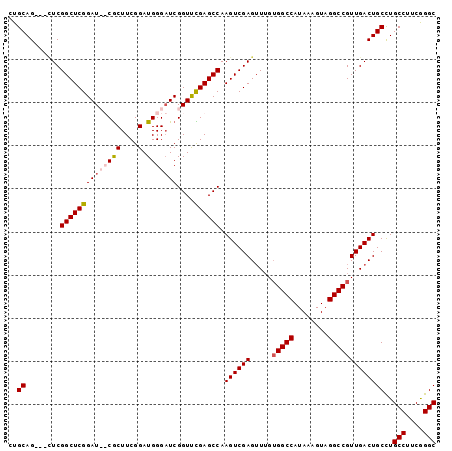
>2R_DroMel_CAF1 4506484 99 - 20766785 CUGCAGCGGCUCGGCUCGGAUCUCGCUUCGGAUGGGAUCGGUUCGAGCCAAGUCGAGUUUGUGGCCAUAAAGUAGGCCGUUGACUGCCUGCCUUCGGGC ..((..(((((.((((((((((((((....).)))))))....)))))).)))))((((..(((((........)))))..))))))..(((....))) ( -46.90) >DroSec_CAF1 40237 96 - 1 CUGCAG---UUCGGCUCGGAUCUCGCUUCGGAUGGGAUCGGUUCGAGCCAAGUCGAGUUUGUGGCCAUAAAGUAGGCCGUUGACUGCCUGCCUUCGGGC ..((((---((.((((((((((((((....).)))))))....)))))).............((((........))))...))))))..(((....))) ( -41.40) >DroEre_CAF1 40175 94 - 1 CUGCAG---CUCGGCUCGGAU--CGCUUCGGAUGGGCUCGGUCUGAGCCAAGUCGAGUUUGUGGCCAUAAAGUAGGCCGUUGACUGCCUGCCUUCGGGC ..((.(---((.(((((((((--((..((.....))..))))))))))).)))..((((..(((((........)))))..))))))..(((....))) ( -41.10) >DroYak_CAF1 42961 94 - 1 CUGCAG---GUCGGCUCGGAU--CACUUCGGAUGGGAUCGGUUUGAGCCAAGUCGAGGUGGGGGCCAUAAAGUAGGCCGUUGACUGCUUGCCUUCAGGC (((.((---((.(((((...(--((((((((...((.((.....)).))...))))))))))))))...((((((........)))))))))).))).. ( -36.40) >consensus CUGCAG___CUCGGCUCGGAU__CGCUUCGGAUGGGAUCGGUUCGAGCCAAGUCGAGUUUGUGGCCAUAAAGUAGGCCGUUGACUGCCUGCCUUCGGGC ..((........((((((((((((((....).)))))))....)))))).((((((.....(((((........)))))))))))))..(((....))) (-34.19 = -35.25 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:04 2006