| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
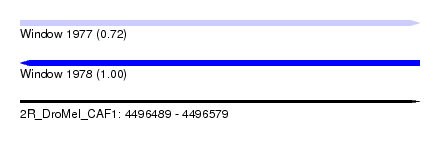
| Location | 4,496,489 – 4,496,579 |
| Length | 90 |
| Max. P | 0.997718 |

| Location | 4,496,489 – 4,496,579 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 87.96 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -18.26 |
| Energy contribution | -19.30 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
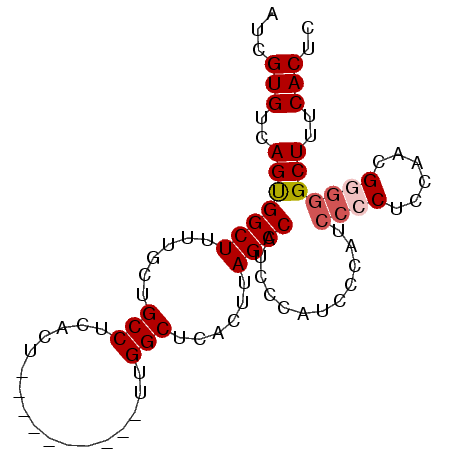
>2R_DroMel_CAF1 4496489 90 + 20766785 GAGUGAAAGCCCCCGUUGGUGGGGAUGGGAUGCGAUGGCUAAGUGAGCCAAAGUGAACCAAAGUGAGGCAGCAAAAGCCACUGACACGAU ..(((..((.(((((....))))).(((..(((..(((((.....)))))..)))..)))......(((.......))).))..)))... ( -27.80) >DroSec_CAF1 29647 79 + 1 GAGUGAAAGCCCCCGUUGGAG-GGAUGGGUUGGGAUGGCUAAGUGAGCCAA----------AGUGAGGCAGCAGAAGCCACUGACACGAU ..(((..(((((((......)-)...))))).((.(((((..((..(((..----------.....))).))...)))))))..)))... ( -22.30) >DroSim_CAF1 29929 80 + 1 GAGUGAAAGCCCCCGUUGGAGGGGAUGAGUUGGGAUGGCUAAGUGAGCCAA----------AGUGAGGCAGCAAAAGCCACUGACACGAU ..((....))((((......))))....((..(..(((((.....))))).----------.....(((.......))).)..))..... ( -24.30) >DroEre_CAF1 29980 75 + 1 GAGUGAAAGCCCCCGAUAGAG-----GGGAUGGGAUGGCUAAGUGAGCCAA----------AGUGAGGCAGCAAAAGCCGCUGACACGAU ..(((..(((((((......)-----))).......((((..((..(((..----------.....))).))...)))))))..)))... ( -25.70) >DroYak_CAF1 32945 80 + 1 GAGUGAAAGCCCCCGAUGGAGGGGAGGGGAUGGGAUGGCUAAGUGAGCCAA----------AGUGAGGCAGCAAAAGCCACUGACACGAU ..(((.....((((......))))......(.((.(((((..((..(((..----------.....))).))...))))))).))))... ( -23.40) >consensus GAGUGAAAGCCCCCGUUGGAGGGGAUGGGAUGGGAUGGCUAAGUGAGCCAA__________AGUGAGGCAGCAAAAGCCACUGACACGAU .((((...(((((((((......))))))......(((((.....)))))................))).((....))))))........ (-18.26 = -19.30 + 1.04)



| Location | 4,496,489 – 4,496,579 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 87.96 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -18.89 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4496489 90 - 20766785 AUCGUGUCAGUGGCUUUUGCUGCCUCACUUUGGUUCACUUUGGCUCACUUAGCCAUCGCAUCCCAUCCCCACCAACGGGGGCUUUCACUC ...(((..((((((.......)))......(((.......(((((.....))))).......))).((((......)))))))..))).. ( -24.44) >DroSec_CAF1 29647 79 - 1 AUCGUGUCAGUGGCUUCUGCUGCCUCACU----------UUGGCUCACUUAGCCAUCCCAACCCAUCC-CUCCAACGGGGGCUUUCACUC ...(((..(((((((......(((.....----------..)))......))))...........(((-(......)))))))..))).. ( -21.30) >DroSim_CAF1 29929 80 - 1 AUCGUGUCAGUGGCUUUUGCUGCCUCACU----------UUGGCUCACUUAGCCAUCCCAACUCAUCCCCUCCAACGGGGGCUUUCACUC ...(((..(((((((......(((.....----------..)))......))))............((((......)))))))..))).. ( -23.90) >DroEre_CAF1 29980 75 - 1 AUCGUGUCAGCGGCUUUUGCUGCCUCACU----------UUGGCUCACUUAGCCAUCCCAUCCC-----CUCUAUCGGGGGCUUUCACUC ...(((..(((((((......(((.....----------..)))......)))).......(((-----(......)))))))..))).. ( -26.20) >DroYak_CAF1 32945 80 - 1 AUCGUGUCAGUGGCUUUUGCUGCCUCACU----------UUGGCUCACUUAGCCAUCCCAUCCCCUCCCCUCCAUCGGGGGCUUUCACUC ...(((..(((((((......(((.....----------..)))......))))............((((......)))))))..))).. ( -23.90) >consensus AUCGUGUCAGUGGCUUUUGCUGCCUCACU__________UUGGCUCACUUAGCCAUCCCAUCCCAUCCCCUCCAACGGGGGCUUUCACUC ...(((..(((((((......(((.................)))......))))............((((......)))))))..))).. (-18.89 = -19.53 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:02 2006