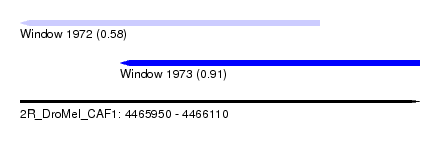
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,465,950 – 4,466,110 |
| Length | 160 |
| Max. P | 0.911449 |

| Location | 4,465,950 – 4,466,070 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -24.95 |
| Energy contribution | -25.73 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4465950 120 - 20766785 ACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAUAGCCACUACCAUAUCCACAUCUACCGCCACCGACACCGCCGAGAGCGUGAUGAAACAA ..(((((.(.((((((..((((.(((((....))))).))))...(((.....)))...............................)))..((....))))).).)))))......... ( -32.70) >DroSec_CAF1 13963 119 - 1 ACCAUGCC-CCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCACAGCCACAUCCAUAUCCACAUCUACCGCCACCGCCACCGCCGAGAGCGUGAUGAAACAA ..((((((-.((((((..((((.(((((....))))).))))...(((.....))).......))).....................((....)).....))).).)))))......... ( -33.20) >DroSim_CAF1 14016 120 - 1 ACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCACAGCCACAUCCAUAUCCACAUCUACCGCCACCGACACCGCCGAGAGCGUGAUGAAACAA ..(((((.(.((((((..((((.(((((....))))).))))...(((.....)))...............................)))..((....))))).).)))))......... ( -32.70) >DroEre_CAF1 14125 108 - 1 ACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAC------------AUCCACAUCUACCGCCACCGACACCGCCGAGAGCGUGAUGAAACAA ..(((((.(.((((((..((((.(((((....))))).))))...(((.....)))......------------.............)))..((....))))).).)))))......... ( -32.70) >DroYak_CAF1 13886 108 - 1 ACCAUGCCCCCGAGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAC------------AUCCACUUCUACCGCCAUCGACACCGCCGAGAGCGUGAUGAAACAA ..........((((((..((((.(((((....))))).))))...(((.....)))......------------.............))).))).((.((((....).))).))...... ( -29.40) >DroAna_CAF1 12961 96 - 1 ACCAUACCCACGGGGCACCUGGCCGGGCACACGCUUACCCAGACAGCCACAGACACAUCCAU------------AAAG------------GCAGACUUUGCCGAAAGCGUGAUGAAACAA ........((((.(((..((((..((((....))))..))))...)))..............------------...(------------((((...))))).....))))......... ( -23.70) >consensus ACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAC____________AUCCACAUCUACCGCCACCGACACCGCCGAGAGCGUGAUGAAACAA ..........((((((..((((.(((((....))))).))))...(((.....)))...............................))).))).((.((((....).))).))...... (-24.95 = -25.73 + 0.78)

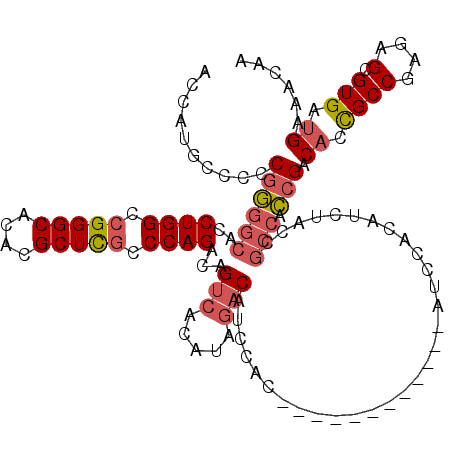
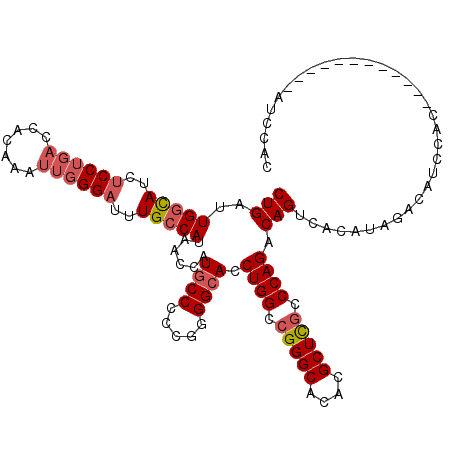
| Location | 4,465,990 – 4,466,110 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.98 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -28.56 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4465990 120 - 20766785 CUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAUAGCCACUACCAUAUCCAC .....(((((..(((..(.......)..)))..)))))......(((((...))))).((((.(((((....))))).))))...(((.....)))........................ ( -36.10) >DroSec_CAF1 14003 119 - 1 CUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCC-CCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCACAGCCACAUCCAUAUCCAC (((..(((((..(((..(.......)..)))..)))))......((((-....)))).((((.(((((....))))).))))...(((.....))).....)))................ ( -36.60) >DroSim_CAF1 14056 120 - 1 CUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCACAGCCACAUCCAUAUCCAC (((..(((((..(((..(.......)..)))..)))))......(((((...))))).((((.(((((....))))).))))...(((.....))).....)))................ ( -36.90) >DroEre_CAF1 14165 108 - 1 CUGAUUGGCAUCUCCUGACCACAAAUUCGGAUUUGCCAUAACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAC------------AUCCAC .....(((((..(((.((........)))))..)))))......(((((...))))).((((.(((((....))))).))))...(((.....)))......------------...... ( -34.00) >DroYak_CAF1 13926 108 - 1 CUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCCCCCGAGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAC------------AUCCAC .....(((((..(((..(.......)..)))..)))))......((((.....)))).((((.(((((....))))).))))...(((.....)))......------------...... ( -33.90) >DroAna_CAF1 12991 96 - 1 CUGAUUGGUAUCUCCU---------UUGGG-GCUGGCAUAACCAUACCCACGGGGCACCUGGCCGGGCACACGCUUACCCAGACAGCCACAGACACAUCCAU------------AAAG-- (((..((((...((((---------.((((-..(((.....)))..)))).))))...((((..((((....))))..))))...)))))))..........------------....-- ( -28.80) >consensus CUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAC____________AUCCAC (((..(((((..((((((.......))))))..)))))......((((.....)))).((((.(((((....))))).)))).))).................................. (-28.56 = -29.45 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:57 2006