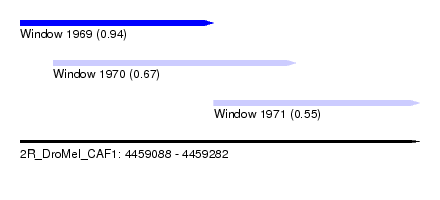
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,459,088 – 4,459,282 |
| Length | 194 |
| Max. P | 0.943562 |

| Location | 4,459,088 – 4,459,182 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 93.86 |
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -29.68 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4459088 94 + 20766785 CAGUC-GAAAGAGUGCUGUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGAGCGGGGGGGUGAGGUGCUUACCCUUCCAGUG ((((.-((..(((((.(((..((((.((((((....))))))))))))).)))))..))))))...(((((((((((...))))))))))).... ( -38.50) >DroSec_CAF1 6745 93 + 1 CAGUC-GAAAGAGUGCUGUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGGGCGGG-GGGUGAGGUGCUUACUCUGCCAGUG ((.((-....)).))...(((((((.((((((((((....(((((....))))))))))))))).)).(-(((((((...))))))))))))).. ( -38.00) >DroSim_CAF1 6814 94 + 1 CAGUC-GAAAGAGUGCUGUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGGGCGGGGGGGUGAGGUGCUUACCCUGCCAGUG ((.((-....)).))...(((((((.((((((((((....(((((....))))))))))))))).))..((((((((...))))))))))))).. ( -41.00) >DroEre_CAF1 7223 94 + 1 CAGUCCGAAAGAGUGCUGUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGGGCGGG-GGGUGAGGAGUGUGCCCUGCCAGUG ((((((....).).))))(((((((.((((((((((....(((((....))))))))))))))).)).(-((((.........)))))))))).. ( -36.10) >DroYak_CAF1 6645 94 + 1 CAGUCCAAAAGAGUGCUGUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUCUAGCGGG-GGGUGAGGAGUUUACCCUGCCAGUG ..........(((((.(((..((((.((((((....))))))))))))).)))))......((.(((((-(.............)))))).)).. ( -28.72) >consensus CAGUC_GAAAGAGUGCUGUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGGGCGGG_GGGUGAGGUGCUUACCCUGCCAGUG ..............((((....(((.((((((((((....(((((....))))))))))))))).)))..(((((((...)))))))...)))). (-29.68 = -30.12 + 0.44)

| Location | 4,459,104 – 4,459,222 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 93.03 |
| Mean single sequence MFE | -50.86 |
| Consensus MFE | -40.90 |
| Energy contribution | -42.26 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4459104 118 + 20766785 GUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGAGCGGGGGGGUGAGGUGCUUACCCUUCCAGUGUGACCCUCGCUCAGCGCUGCGGAAAUCCGUGCAGCUGUGG (((((....)))))(((((((((((.(((((.((((((..((.......))..)))))).))))).......)))))))))))....(.((((((.((((....)))))).)))).). ( -48.50) >DroSec_CAF1 6761 117 + 1 GUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGGGCGGG-GGGUGAGGUGCUUACUCUGCCAGUGUGACCCUCGCGCGGCGCUGCGGAAAUCCGUGCAGCUGUGG (((((....)))))(((((((((((((((((.((((((..((.......)).-)))))).)))))......))))))))))))....((((((((.((((....)))))).)))))). ( -51.70) >DroSim_CAF1 6830 118 + 1 GUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGGGCGGGGGGGUGAGGUGCUUACCCUGCCAGUGUGACCCUCGCGCGGCGCUGCGCAAAUCCGUGCAGCUGUGG ....((((.((((((((((....(((((....))))))))))))))).)((.((((((((...)))))))).)).(((((.....))))))))(((((((......)))))))..... ( -58.00) >DroEre_CAF1 7240 116 + 1 GUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGGGCGGG-GGGUGAGGAGUGUGCCCUGCCAGUGUGACCCUCGCGGGGAGCUGCCGAAAUCUG-GCAGCUGUGG .(((((((.((((((((((....(((((....))))))))))))))).)).(-((((.........)))))))))).....(((.....)))((((((((.....))-)))))).... ( -49.70) >DroYak_CAF1 6662 117 + 1 GUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUCUAGCGGG-GGGUGAGGAGUUUACCCUGCCAGUGUGACCCUCGCGCGGCGCUGCGGAAAUCUGUGCAGCUGUGG ....((((((....(((((((((((..(......((((..((((((......-)))))).)))).....)..)))))))))))...)).))))((((((.(....).))))))..... ( -46.40) >consensus GUUGGCCGGCCAAUGGUUAUAUUGGUGGCACGUCACUUGAUCAUUGGGCGGG_GGGUGAGGUGCUUACCCUGCCAGUGUGACCCUCGCGCGGCGCUGCGGAAAUCCGUGCAGCUGUGG ....((((((....(((((((((((((((((.((((((..((.......))..)))))).)))))......))))))))))))...)).))))((((((........))))))..... (-40.90 = -42.26 + 1.36)

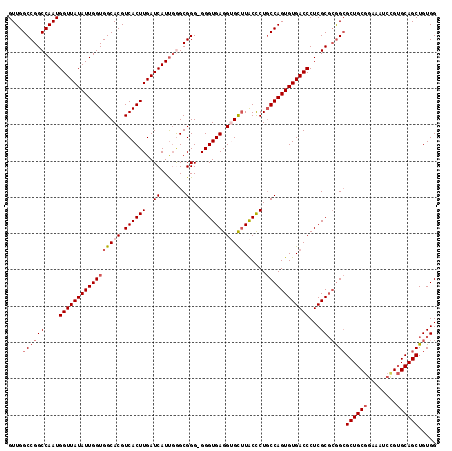
| Location | 4,459,182 – 4,459,282 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -25.99 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4459182 100 + 20766785 UGACCCUCGCUCAGCGCUGCGGAAAUCCGUGCAGCUGUGGUGCCGCCGCUGCGCAAUACUUUUGUCACAUGUGGCGCAAAAUUCAAAUGCGG-A--------------UUUUUGA .............((((..(((....))(((((((.(((....))).)))))))................)..))))(((((((......))-)--------------))))... ( -34.70) >DroSec_CAF1 6838 114 + 1 UGACCCUCGCGCGGCGCUGCGGAAAUCCGUGCAGCUGUGGUGCUGGCGCUGCGCAAUACUUUUGCCACAUGUGGCGCGAAAUUCAAAUGCGG-AUGCAGACAGACGAAUUUUUCA ........(((((((((.((((....)))).((((......))))))))))))).........((((....))))(.(((((((...(((..-..))).......))))))).). ( -43.00) >DroSim_CAF1 6908 113 + 1 UGACCCUCGCGCGGCGCUGCGCAAAUCCGUGCAGCUGUGGUGCUGGCGCUGCGCAAUACUUUUGCCACAUGUGGCGCAAAAUUCAAAUGCGG-AUGCAGA-AGACGAAUUUUUCA ........((((((((((((((......))))(((......))))))))))))).........((((....))))(.(((((((...(((..-..)))..-....))))))).). ( -41.40) >DroEre_CAF1 7317 107 + 1 UGACCCUCGCGGGGAGCUGCCGAAAUCUG-GCAGCUGUGGUGGUGCCGCCUCGCAAUAAUUUUGCCACAUUUGGCGCAAUAUUCAAAGGCGGUCCGCAAGAAG-------UUUCC ...(((.....)))((((((((.....))-))))))...((((.(((((((...((((.((.(((((....))))).))))))...)))))))))))......-------..... ( -42.50) >consensus UGACCCUCGCGCGGCGCUGCGGAAAUCCGUGCAGCUGUGGUGCUGCCGCUGCGCAAUACUUUUGCCACAUGUGGCGCAAAAUUCAAAUGCGG_AUGCAGA_AG_____UUUUUCA ....((.((((((((((((((........))))))...((.....))))))))).........((((....)))).............).))....................... (-25.99 = -26.30 + 0.31)

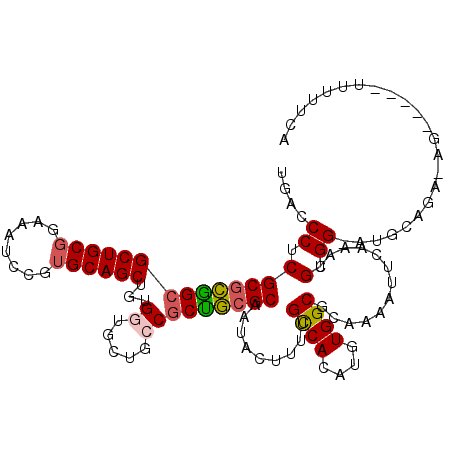
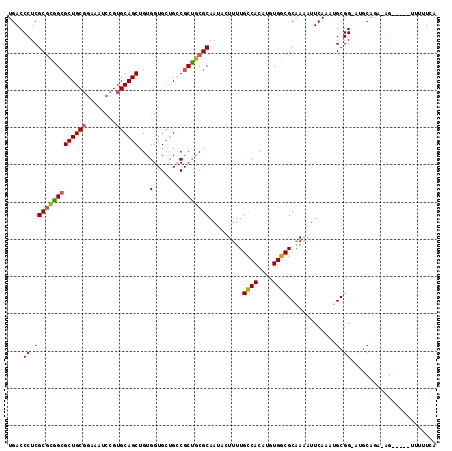
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:55 2006