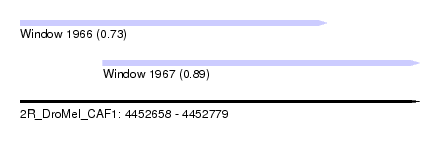
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,452,658 – 4,452,779 |
| Length | 121 |
| Max. P | 0.891274 |

| Location | 4,452,658 – 4,452,751 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.15 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -19.11 |
| Energy contribution | -18.89 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4452658 93 + 20766785 -----UAGCGAUCGGUCC----UCCGCGACCGAUGUGUUGCCAAAGGCGGCUGCUU--CCC-CUCGAGCUCUGCAAUCCAGC-GAACCAAAGGCAGCAUUC---UCCAC -----.....(((((((.----.....)))))))((((((((....((((..((((--...-...)))).))))..((....-))......))))))))..---..... ( -29.30) >DroPse_CAF1 29308 102 + 1 GCUACUGCUGAUCGG-CC----AAUAUGGUCGAUGUGUUGCCAAAGGCGCCUGCUU--UUCUUUCGAGCUCUGCAAAGUAGCAGAACCAAAGGCAGCAUUUAAUUUAAC ((....))..(((((-((----.....)))))))((((((((((((((....))))--)).........(((((......)))))......)))))))).......... ( -33.30) >DroEre_CAF1 847 93 + 1 -----UAGCGAUCGGUCC----UACACGGCCGAUGUGUUGCCAAAGGCGGCUGCUU--UCC-UUCGAGCUCUGCAACCCAGC-GAACCAAAGGCAGCAUUG---UCCAC -----.(((((((((((.----.....))))))).))))(((...))).(((((((--(..-((((....(((.....))))-)))...))))))))....---..... ( -27.80) >DroYak_CAF1 419 93 + 1 -----UAGCGAUCGGACC----UCCACGGCCGAUGUGUUGCCAAAGGCGGAUGCUU--UUC-UUCGAGCUCUGCAAUUCAGC-GAACCAAAGGCAGCAUUC---UCCAC -----.....(((((.((----.....)))))))((((((((....(((((.((((--...-...)))))))))..(((...-))).....))))))))..---..... ( -29.70) >DroAna_CAF1 758 95 + 1 --------AGAUCGGAGUCCCAUUUAUGACCGAUGUGUUGCCAAAGGCGCCUGCUUUUUUC-UUCGA-UUCUGCAAAGUAGC-GAACCAAAGGCAGCAUAA---ACAAU --------..(((((..((........)))))))((((((((((((((....))))))...-((((.-..((....))...)-))).....))))))))..---..... ( -24.00) >DroPer_CAF1 30236 102 + 1 GCUACUGCUGAUCGG-CC----AAUAUGGUCGAUGUGUUGCCAAAGGCGCCUGCUU--UUCUUUCGAGCUCUGCAAAGUAGCAGAACCAAAGGCAGCAUUUAAUUUAAC ((....))..(((((-((----.....)))))))((((((((((((((....))))--)).........(((((......)))))......)))))))).......... ( -33.30) >consensus _____UAGCGAUCGG_CC____UACACGGCCGAUGUGUUGCCAAAGGCGCCUGCUU__UUC_UUCGAGCUCUGCAAAGCAGC_GAACCAAAGGCAGCAUUC___UCAAC ..........(((((..............)))))((((((((..((((....)))).............((.((......)).))......)))))))).......... (-19.11 = -18.89 + -0.22)


| Location | 4,452,683 – 4,452,779 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -15.27 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4452683 96 + 20766785 GUGUUGCCAAAGGCGGCUGCUU--CCC-CUCGAGCUCUGCAAUCCAGC-GAACCAAAGGCAGCAUUC---UCCACA---CCCAAACUC--ACACACAAACAUCACAAU ((((((((....((((..((((--...-...)))).))))..((....-))......))))))))..---......---.........--.................. ( -19.20) >DroPse_CAF1 29337 97 + 1 GUGUUGCCAAAGGCGCCUGCUU--UUCUUUCGAGCUCUGCAAAGUAGCAGAACCAAAGGCAGCAUUUAAUUUAACC---CACAUACAC--AUACA----CAUCAUAAU ((((((((((((((....))))--)).........(((((......)))))......))))))))...........---.........--.....----......... ( -22.10) >DroEre_CAF1 872 92 + 1 GUGUUGCCAAAGGCGGCUGCUU--UCC-UUCGAGCUCUGCAACCCAGC-GAACCAAAGGCAGCAUUG---UCCACU---CCUAAACUC--ACGCAC----AUCACAAU ((((((((...))))(((((((--(..-((((....(((.....))))-)))...))))))))....---......---.........--..))))----........ ( -20.70) >DroYak_CAF1 444 89 + 1 GUGUUGCCAAAGGCGGAUGCUU--UUC-UUCGAGCUCUGCAAUUCAGC-GAACCAAAGGCAGCAUUC---UCCACU---CCUAAACUC--ACGCAC----AUCA---U ((((((((....(((((.((((--...-...)))))))))..(((...-))).....))))))))..---......---.........--......----....---. ( -21.00) >DroAna_CAF1 784 98 + 1 GUGUUGCCAAAGGCGCCUGCUUUUUUC-UUCGA-UUCUGCAAAGUAGC-GAACCAAAGGCAGCAUAA---ACAAUGUACUUUAAACUUUCACCCA----CAUCACAAU ((((((((((((((....))))))...-((((.-..((....))...)-))).....))))))))..---.........................----......... ( -19.70) >DroPer_CAF1 30265 97 + 1 GUGUUGCCAAAGGCGCCUGCUU--UUCUUUCGAGCUCUGCAAAGUAGCAGAACCAAAGGCAGCAUUUAAUUUAACC---CACAUACAC--AUACA----CAUCAUAAU ((((((((((((((....))))--)).........(((((......)))))......))))))))...........---.........--.....----......... ( -22.10) >consensus GUGUUGCCAAAGGCGCCUGCUU__UUC_UUCGAGCUCUGCAAAGCAGC_GAACCAAAGGCAGCAUUC___UCAACC___CCCAAACUC__ACACA____CAUCACAAU ((((((((..((((....)))).............((.((......)).))......))))))))........................................... (-15.27 = -15.27 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:51 2006