| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,442,220 – 4,442,319 |
| Length | 99 |
| Max. P | 0.988555 |

| Location | 4,442,220 – 4,442,319 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -13.26 |
| Energy contribution | -13.07 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
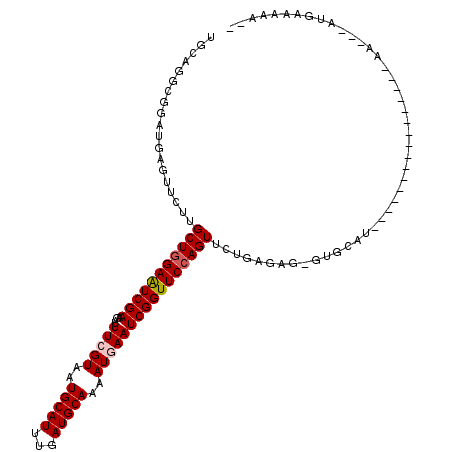
>2R_DroMel_CAF1 4442220 99 + 20766785 GAUAUUUAAU---UU----------------AUGCACACUCUCAGAACUGGAACCGAUUCAUUUUGCAUCAAAUGCAUUACGAAAUAUUCGAUUCUAGCAAGAACGCAUCCACCUACA ..........---..----------------((((....(((.....((((((.((((((.(..(((((...)))))..).)))....))).))))))..)))..))))......... ( -14.30) >DroPse_CAF1 5421 114 + 1 --AUUAUCAUAAACUUUGGAGACAAACAUCUCUGCA--UCUACAGAACUGGAACCGAUUUAUACUGCAUCAAAUGCAUUACGAAAUCUUCGAUUCCAGCAAGAACUCAUCUGUGGGAG --...............(((((......)))))...--((((((((.((((((.(((((..((.(((((...))))).))..))....))).))))))...(....).)))))))).. ( -26.60) >DroEre_CAF1 7570 99 + 1 GAUAUUUAAU---UU----------------AUGCACUCUCUUAGAACUGGAACCGAUUCAUUUUGCAUCAAAUGCAUUACGAAAUUUUCGAUUCCAGCAAGAACCCAUACACCUUCA ..........---.(----------------(((.....((((....((((((.((((((.(..(((((...)))))..).)))....))).)))))).))))...))))........ ( -15.90) >DroYak_CAF1 7426 99 + 1 GAUGUUUAAU---UU----------------AUGCACACUCUCAGAACUGGAACCGAUUCAUUUUGCAUCAAAUGCAUUACGAAAUUUUCGAUUCCAGCAAGAACACAUCCACCUCCA (((((....(---((----------------.(((...(.....)...(((((.((((((.(..(((((...)))))..).)))....))).)))))))).))).)))))........ ( -17.00) >DroAna_CAF1 7320 108 + 1 --UUUUUCAU---UUUUGCUAAUACA-----AUGCACGCCAUUAGAACUGGAACCGAUUCAUUCUGCAUCAAAUGCAUUACGAAAUCUUCGACUCCAGCAAGAACUCGUCCGCUUCAA --.......(---(((((((.....(-----(((((......(((((.((((.....))))))))).......)))))).(((.....))).....)))))))).............. ( -17.32) >DroPer_CAF1 5470 114 + 1 --AUUAUCAUAAACUUUGGAGACAAACAUCUCUGCA--UCUACAGAACUGGAACCGAUUUAUACUGCAUCAAAUGCAUUACGAAAUCUUCGAUUCCAGCAAGAACUCAUCUGUGGGAG --...............(((((......)))))...--((((((((.((((((.(((((..((.(((((...))))).))..))....))).))))))...(....).)))))))).. ( -26.60) >consensus __UUUUUAAU___UU________________AUGCAC_CCCUCAGAACUGGAACCGAUUCAUUCUGCAUCAAAUGCAUUACGAAAUCUUCGAUUCCAGCAAGAACUCAUCCACCUCAA ...............................................((((((.((((((.(..(((((...)))))..).)))....))).)))))).................... (-13.26 = -13.07 + -0.19)


| Location | 4,442,220 – 4,442,319 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4442220 99 - 20766785 UGUAGGUGGAUGCGUUCUUGCUAGAAUCGAAUAUUUCGUAAUGCAUUUGAUGCAAAAUGAAUCGGUUCCAGUUCUGAGAGUGUGCAU----------------AA---AUUAAAUAUC .....(((.((((.(((..(((.(((((((....(((((..(((((...)))))..)))))))))))).)))...))).)))).)))----------------..---.......... ( -24.70) >DroPse_CAF1 5421 114 - 1 CUCCCACAGAUGAGUUCUUGCUGGAAUCGAAGAUUUCGUAAUGCAUUUGAUGCAGUAUAAAUCGGUUCCAGUUCUGUAGA--UGCAGAGAUGUUUGUCUCCAAAGUUUAUGAUAAU-- .....(((((.........(((((((((((....((..((.(((((...))))).))..))))))))))))))))))(((--(...(((((....)))))....))))........-- ( -27.80) >DroEre_CAF1 7570 99 - 1 UGAAGGUGUAUGGGUUCUUGCUGGAAUCGAAAAUUUCGUAAUGCAUUUGAUGCAAAAUGAAUCGGUUCCAGUUCUAAGAGAGUGCAU----------------AA---AUUAAAUAUC .....((((((...((((((((((((((((....(((((..(((((...)))))..))))))))))))))))...))))).))))))----------------..---.......... ( -31.10) >DroYak_CAF1 7426 99 - 1 UGGAGGUGGAUGUGUUCUUGCUGGAAUCGAAAAUUUCGUAAUGCAUUUGAUGCAAAAUGAAUCGGUUCCAGUUCUGAGAGUGUGCAU----------------AA---AUUAAACAUC .....(((.(..(.(((..(((((((((((....(((((..(((((...)))))..))))))))))))))))...))).)..).)))----------------..---.......... ( -29.10) >DroAna_CAF1 7320 108 - 1 UUGAAGCGGACGAGUUCUUGCUGGAGUCGAAGAUUUCGUAAUGCAUUUGAUGCAGAAUGAAUCGGUUCCAGUUCUAAUGGCGUGCAU-----UGUAUUAGCAAAA---AUGAAAAA-- .......(.(((.(((...((((((..(((....(((((..(((((...)))))..))))))))..))))))...)))..))).).(-----(((....))))..---........-- ( -28.80) >DroPer_CAF1 5470 114 - 1 CUCCCACAGAUGAGUUCUUGCUGGAAUCGAAGAUUUCGUAAUGCAUUUGAUGCAGUAUAAAUCGGUUCCAGUUCUGUAGA--UGCAGAGAUGUUUGUCUCCAAAGUUUAUGAUAAU-- .....(((((.........(((((((((((....((..((.(((((...))))).))..))))))))))))))))))(((--(...(((((....)))))....))))........-- ( -27.80) >consensus UGCAGGCGGAUGAGUUCUUGCUGGAAUCGAAGAUUUCGUAAUGCAUUUGAUGCAAAAUGAAUCGGUUCCAGUUCUGAGAG_GUGCAU________________AA___AUGAAAAA__ ...................(((((((((((....(((((..(((((...)))))..)))))))))))))))).............................................. (-19.45 = -19.82 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:48 2006