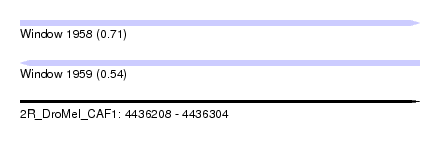
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,436,208 – 4,436,304 |
| Length | 96 |
| Max. P | 0.712761 |

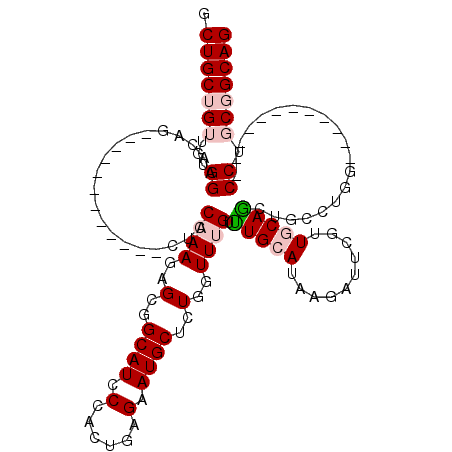
| Location | 4,436,208 – 4,436,304 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -18.94 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4436208 96 + 20766785 CUGCCGCAGG------------CCAGGCAGCUGCAACAAAUCUUAUGCAAACGAACCAGAGCAUUCUCAGUGGGAUGCCGCUCUUUGGAG------------CUGUUCCGCAACAGCAGC .((((.....------------...))))(((((........................(((....))).(((((((...((((....)))------------).)))))))....))))) ( -29.60) >DroPse_CAF1 6878 120 + 1 CUGCAGGCGGAAGCUUGCCCGUGCAAGGAUCUGGAUCGAAUCUGAUGCAGACAAAUCAAAGCAUUCUUAGUGGGAUGCCCCUGUUUGGGGCACACCCACAAGCGGUUCCACAGCAGCAGC ..((((((....))))))((((((...((((((.((((....)))).))))....))...)).......(((((.((((((.....))))))..)))))..))))............... ( -48.10) >DroSec_CAF1 6059 96 + 1 CUGCCGCAGG------------CCAGGCAGUUGCAACGAAUCUUAUGCAAACAAACCAGAGCAUCCUCAGUGGGAUGCCGCUCUUUGGAG------------CUGUUCCACAACAGCAGC (((((.....------------...)))))(((((..........))))).....((((((((((((....))))))).....))))).(------------(((((....))))))... ( -31.20) >DroSim_CAF1 6573 96 + 1 CUGCCGCAGG------------CCAGGCAGCUGCAACGAAUCUUAUGCAAACGAACCAGAGCAUCCUCAGUGGGAUGCCGCUCUUUGGAG------------CUGUUCCACAACAGCAGC .((((.....------------...))))(((((........................(((....))).(((((((...((((....)))------------).)))))))....))))) ( -30.20) >DroPer_CAF1 6893 120 + 1 CUGCAGGCGGAACCUUGCCCGUGCAAGGAUCUGGAUCGAAUCUGAUGCAGACAAAUCAAAGCAUUCUUAGUGGGAUGCCCCUGUUUGGGGCACACCCACAAGCGGUUCCACAGCAGCAGC ((((..(.((((((.(((((......)).((((.((((....)))).)))).........)))......(((((.((((((.....))))))..)))))....)))))).).)))).... ( -48.90) >consensus CUGCCGCAGG____________CCAGGCAGCUGCAACGAAUCUUAUGCAAACAAACCAGAGCAUUCUCAGUGGGAUGCCGCUCUUUGGAG____________CUGUUCCACAACAGCAGC ((((.....................(((..(((((..........)))))..........(((((((....))))))).)))...(((((...............))))).....)))). (-18.94 = -18.22 + -0.72)

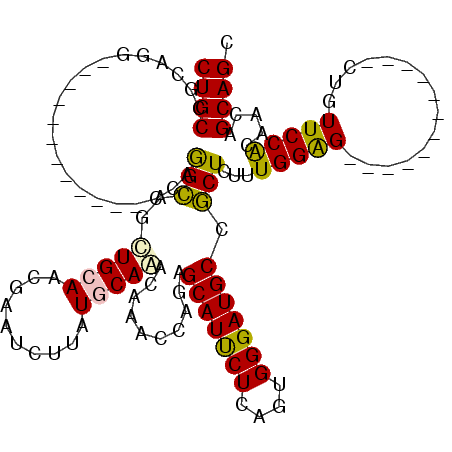
| Location | 4,436,208 – 4,436,304 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -13.14 |
| Energy contribution | -14.34 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4436208 96 - 20766785 GCUGCUGUUGCGGAACAG------------CUCCAAAGAGCGGCAUCCCACUGAGAAUGCUCUGGUUCGUUUGCAUAAGAUUUGUUGCAGCUGCCUGG------------CCUGCGGCAG ....(((((((((..(((------------(((....))))((((..(((..(((....)))))).....(((((..........))))).)))))).------------.))))))))) ( -31.70) >DroPse_CAF1 6878 120 - 1 GCUGCUGCUGUGGAACCGCUUGUGGGUGUGCCCCAAACAGGGGCAUCCCACUAAGAAUGCUUUGAUUUGUCUGCAUCAGAUUCGAUCCAGAUCCUUGCACGGGCAAGCUUCCGCCUGCAG ....((((.((((((..(((((((((.(((((((.....)))))))))))).......((((((....(((((.(((......))).))))).....)).))))))))))))))..)))) ( -49.10) >DroSec_CAF1 6059 96 - 1 GCUGCUGUUGUGGAACAG------------CUCCAAAGAGCGGCAUCCCACUGAGGAUGCUCUGGUUUGUUUGCAUAAGAUUCGUUGCAACUGCCUGG------------CCUGCGGCAG ....((((((..(..(((------------(((....))))(((((((......)))))))..(((..(((.(((..........)))))).))))).------------.)..)))))) ( -33.10) >DroSim_CAF1 6573 96 - 1 GCUGCUGUUGUGGAACAG------------CUCCAAAGAGCGGCAUCCCACUGAGGAUGCUCUGGUUCGUUUGCAUAAGAUUCGUUGCAGCUGCCUGG------------CCUGCGGCAG .(((((((.((.(..(((------------(......(((((((((((......)))))))...))))...((((..........))))))))..).)------------)..))))))) ( -33.70) >DroPer_CAF1 6893 120 - 1 GCUGCUGCUGUGGAACCGCUUGUGGGUGUGCCCCAAACAGGGGCAUCCCACUAAGAAUGCUUUGAUUUGUCUGCAUCAGAUUCGAUCCAGAUCCUUGCACGGGCAAGGUUCCGCCUGCAG ....((((.((((((((.((((((((.(((((((.....)))))))))))).)))..(((((((....(((((.(((......))).))))).....)).))))).))))))))..)))) ( -55.30) >consensus GCUGCUGUUGUGGAACAG____________CUCCAAAGAGCGGCAUCCCACUGAGAAUGCUCUGGUUUGUUUGCAUAAGAUUCGUUGCAGCUGCCUGG____________CCUGCGGCAG .(((((((...((....................((((..(..((((.(......).))))..)..)))).(((((..........)))))....................)).))))))) (-13.14 = -14.34 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:44 2006