
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,427,164 – 4,427,263 |
| Length | 99 |
| Max. P | 0.881811 |

| Location | 4,427,164 – 4,427,263 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.77 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4427164 99 + 20766785 GCUGUCAACGCCAAGCACCGCCAACAACUCCCCCACCACCACGCCCUGUAGUUCCCGGGCGGCGUCGUACGCCCAGCUGCU---------------------GCACAACAUCUUGCCACA (((..........)))..............................((((((..(.(((((........))))).)..)))---------------------)))............... ( -20.20) >DroVir_CAF1 3162 96 + 1 CUUGU------CCAGCAGCACAAACAAUUCACCCACAAAUACGCCAUGCAAUUCGCGUGCCGCUUCCUAUGCGCAGCUGCUG------------------GUCAACAAUAUCAUGCCACA .(((.------((((((((.........................(((((.....))))).(((.......)))..)))))))------------------).)))............... ( -27.10) >DroPse_CAF1 4032 117 + 1 GCUGUCAACACCCAGCAGCACCAACAACUCACCCACAACGACGCCAUGCAGCUCGAGGGCAACCUCCUAUGCGCAGCUGCU---CAGUGUACAGAGCUUAGUCCACAAUAUCUUGCCACA .((((..((((..((((((....................(....).(((.((..((((....))))....)))))))))))---..)))))))).......................... ( -29.70) >DroEre_CAF1 3370 99 + 1 GCUGUCAACGCCAAGCACCGCCAACAACUCACCCACCACCACGCCCUGCAGCUCCCGGGCGGCAUCCUACGCCCAGCUUCU---------------------GCACAAUAUCUUGCCACA (((..........)))..............................(((((...(.(((((........))))).)...))---------------------)))............... ( -18.70) >DroMoj_CAF1 3424 114 + 1 GCUGC------CCAGCAGCACAAACAACUCGCCGACAACGACGCCCAGUAAUUCGCGUGCCGCCUCCUAUGCGCAACUACUGGGCGGUGUCCAGCACUUAGUCAACAAUAUCUUGCCACA (((((------...)))))...........((.((((....(((((((((.((((((((........)))))).)).))))))))).))))..))......................... ( -37.20) >DroPer_CAF1 4104 117 + 1 GCUGUCAACACCCAGCAGCACCAACAACUCACCCACAACGACGCCAUGCAGCUCGAGGGCAACCUCCUAUGCGCAGCUGCU---CAGUGUGCAGAGCUUAGUCCACAAUAUCUUGCCACA (((((.........)))))....................(((((...))(((((((((....))))...((((((.((...---.)))))))))))))..)))................. ( -31.90) >consensus GCUGUCAAC_CCCAGCAGCACCAACAACUCACCCACAACGACGCCAUGCAGCUCGCGGGCAGCCUCCUAUGCGCAGCUGCU___________________GUCCACAAUAUCUUGCCACA (((((.........)))))............................((((((...(((((........)))))))))))........................................ (-12.40 = -12.73 + 0.34)
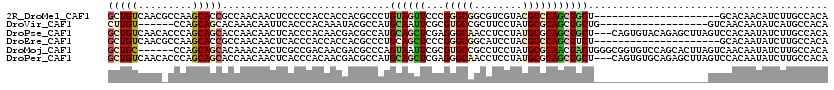

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:41 2006