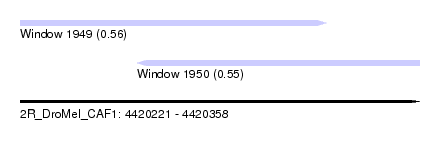
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,420,221 – 4,420,358 |
| Length | 137 |
| Max. P | 0.556839 |

| Location | 4,420,221 – 4,420,326 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -18.29 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
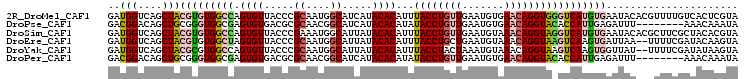
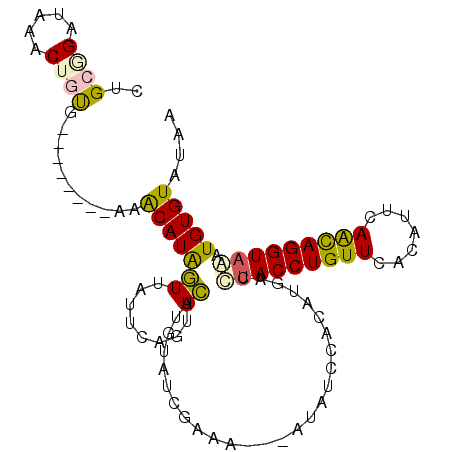
>2R_DroMel_CAF1 4420221 105 + 20766785 GAUGGUCAGCUACGUGUGGCCAGUGUUACCCGCAAUGGCAUCAUACACAUUUACCUGUUGAAUGUGAACAGGUGGGUCAUGUGAAUACACGUUUUGUCACUCGUA (((((.(((..(((((((((((.(((.....))).)))).(((((...(((((((((((.......)))))))))))..))))).))))))).)))))).))... ( -34.40) >DroPse_CAF1 2512 97 + 1 GACGGACAGCUGCGGGUGGCGAGUGUGACGCGCAACGGCAUCAUACACAUAUACCUGUUGAAUGUGAACAGGUACACCAUUGAGAUUU--------AAACAAAUA (.(((....)))).((((....((((((.((......)).)))))).....((((((((.......))))))))))))..........--------......... ( -25.70) >DroSim_CAF1 2723 105 + 1 GAUGGUCAGCUACGUGUGGCCAGUGUUACCCGAAAUGGCAUUAUACACAUUUACCUGUUGAAUGUAAACAGGUAGGUCAUGUGAAUACACGCUUCGCUACACGUA ..........((((((((((.(((((..((......)).......((((((((((((((.......)))))))))...))))).....)))))..)))))))))) ( -34.00) >DroEre_CAF1 2705 103 + 1 GAUGGUCAGCUACGUGUGGCUAGUGUUACCCGCAAUGGCAUUAUACACAUUUACCUGCUGAAUGUAAACAGGUAAGUCAAGUGAUUAA--UUUUCGAUACAAGUA ..((((((.((..(((((((((.(((.....))).))))....)))))(((((((((...........)))))))))..)))))))).--............... ( -22.00) >DroYak_CAF1 2959 103 + 1 GAUGGUCAGCUACGCGUGGCCAGUGUUACCCGCAAUGGCAUUAUACACAUUUACCUACUAAAUGUAAACAGGUAAGUCAAGUGGUUAU--UUUUCGAUAUAAGUA ((..(..((((((...((((((.(((.....))).)))).........((((((((((.....))....)))))))))).))))))..--)..)).......... ( -21.40) >DroPer_CAF1 2511 97 + 1 GACGGACAGCUGCGGGUGGCGAGUGUGACGCGCAACGGCAUCAUACACAUAUACCUGUUGAAUGUGAACAGGUACACCAUUGAGAUUU--------AAACAAAUA (.(((....)))).((((....((((((.((......)).)))))).....((((((((.......))))))))))))..........--------......... ( -25.70) >consensus GAUGGUCAGCUACGUGUGGCCAGUGUUACCCGCAAUGGCAUCAUACACAUUUACCUGUUGAAUGUAAACAGGUAAGUCAUGUGAAUAU___UUUCGAUACAAGUA ..(((....)))(((((((((.((((.....((....)).....))))...((((((((.......)))))))))))))))))...................... (-18.29 = -18.68 + 0.39)
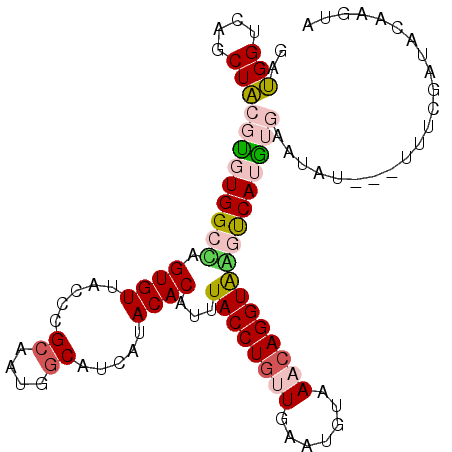
| Location | 4,420,261 – 4,420,358 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.44 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -10.27 |
| Energy contribution | -9.67 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4420261 97 - 20766785 CUGCGGAUAAACUGUG--------AAACAUAGUACUUCAGUACGAGUGACAAAACGUGUAUUCACAUGACCCACCUGUUCACAUUCAACAGGUAAAUGUGUAUGA .((((.((....((((--------((((((.((((....))))..((......)))))).))))))......(((((((.......)))))))..)).))))... ( -22.40) >DroPse_CAF1 2552 97 - 1 CGGCUGAUAGACUACGGAUAGUAUAAGCAUGAUUAUGUAUUAUUUGUUU--------AAAUCUCAAUGGUGUACCUGUUCACAUUCAACAGGUAUAUGUGUAUGA ......(((.((.((((((((((((..........))))))))))))..--------...........(((((((((((.......))))))))))))).))).. ( -22.00) >DroSim_CAF1 2763 98 - 1 CUGCGGAUAAACUGUGG-------AGACAUAGUUAUUCAGUACGUGUAGCGAAGCGUGUAUUCACAUGACCUACCUGUUUACAUUCAACAGGUAAAUGUGUAUAA ....((((((.(((((.-------...)))))))))))..(((((........)))))(((.(((((....((((((((.......)))))))).))))).))). ( -30.50) >DroEre_CAF1 2745 95 - 1 CUGCAGAUAAACUGUG--------AAACAUAGUUCUUCAGUACUUGUAUCGAAAA--UUAAUCACUUGACUUACCUGUUUACAUUCAGCAGGUAAAUGUGUAUAA .((((((..(((((((--------...)))))))..)).................--.............(((((((((.......)))))))))...))))... ( -18.40) >DroYak_CAF1 2999 95 - 1 CUGCGGAUAAACUGUA--------AAACAUAGUUUUUCAGUACUUAUAUCGAAAA--AUAACCACUUGACUUACCUGUUUACAUUUAGUAGGUAAAUGUGUAUAA .(((((.....)))))--------..((((((((((((.(((....))).)))))--))...........(((((((((.......))))))))).))))).... ( -15.50) >DroPer_CAF1 2551 97 - 1 CGGCUGAUAGACUACGGAUAGUAUAAGCAUGAUUAUGUAUUAUUUGUUU--------AAAUCUCAAUGGUGUACCUGUUCACAUUCAACAGGUAUAUGUGUAUGA ......(((.((.((((((((((((..........))))))))))))..--------...........(((((((((((.......))))))))))))).))).. ( -22.00) >consensus CUGCGGAUAAACUGUG________AAACAUAGUUAUUCAGUACUUGUAUCGAAA___AUAUCCACAUGACCUACCUGUUCACAUUCAACAGGUAAAUGUGUAUAA ..((((.....))))...........(((((((........))...........................(((((((((.......))))))))).))))).... (-10.27 = -9.67 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:35 2006