| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,414,260 – 4,414,402 |
| Length | 142 |
| Max. P | 0.993078 |

| Location | 4,414,260 – 4,414,380 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -26.99 |
| Energy contribution | -28.10 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4414260 120 + 20766785 CUAAGAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGACUACCUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAGUCGUCCUGCACCGUUCUCUAAGGAGGCG ......(((((.....))))).((((.(((.((((((.((((.(.(((.....(((((((((........))))))))).....))).).)))).))))))..))).))))......... ( -46.00) >DroVir_CAF1 1352 120 + 1 ACAAAAUGAAGAAAUAUAUAUCGGAGCACGAGCAGGAGGAUUACCUAUUGACUGGUUUCACCAGCCAGAAGGUAAAUCCCUUCCGCGAGAAAUCCUCGUGCACUGCUCUUUAAGCCGAUU ...................(((((.....((((((..(((((((((.(((.((((.....)))).))).)))).)))))....((((((.....))))))..))))))......))))). ( -41.90) >DroGri_CAF1 1780 120 + 1 AUAAAAUGAAGAAAUAUAUAUCGGAGCACGAGCAGGAGGAUUAUCUAUUGACUGGUUUCACCAGCCAAAAGGUAAACCCCUUCCGUGAGAAGUCCUCGUGCACGGCUCUCUAAGCAGAUC ......................(((((.((.(((.(((((((((((.(((.((((.....)))).))).))))))....(((.(....))))))))).))).)))))))........... ( -38.40) >DroWil_CAF1 1784 120 + 1 ACAAAAUGAUGAAAUAUAUCACAGAGCAUGAGCAGGAGGAUUAUCUAUUGACUGGUUUCACCAGCCAAAAGGUUAAUCCGUUCCGUGAGAAAUCCUCAUGCACAGUGCUUUAAGGCGGUC ......(((((.....))))).(((((((..((.((((((((((((.(((.((((.....)))).))).))).))))))..)))(((((.....)))))))...)))))))......... ( -34.70) >DroMoj_CAF1 1696 120 + 1 AUAAAAUGAAGAAGUAUAUAUCGGAGCACGAGCAGGAGGAUUAUCUGUUGACUGGCUUCACCAGUCAGAAGGUAAACCCAUUCCGUGAGAAGUCGUCGUGCACUGCUCUUUAAGCAGGUU .........................((((((...(((((.((((((.((((((((.....)))))))).)))))).))...)))..((....)).)))))).(((((.....)))))... ( -37.00) >DroAna_CAF1 1946 120 + 1 CUAAAAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGACUACCUGCUGACUGGAUUCACCAGCCAGAAGGUUAAUCCCUUCCGCGAGAAGUCAUCAUGCACCGUACUUUAAGAUGGUG .....(((((((..(...((.((((((....)).((.(((..((((.(((.((((.....)))).))).))))...))))))))).)).)..))))))).((((((........)))))) ( -40.60) >consensus AUAAAAUGAAGAAAUAUAUAACGGAGCACGAGCAGGAGGAUUACCUAUUGACUGGUUUCACCAGCCAGAAGGUAAAUCCCUUCCGCGAGAAGUCCUCGUGCACCGCUCUUUAAGCAGGUC .....................(((.((((((((.((((((((((((.(((.((((.....)))).))).)))).)))))..)))))((....)).)))))).)))............... (-26.99 = -28.10 + 1.11)

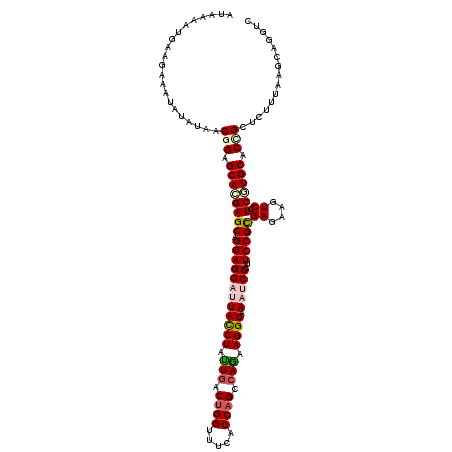
| Location | 4,414,260 – 4,414,380 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -41.18 |
| Consensus MFE | -27.07 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4414260 120 - 20766785 CGCCUCCUUAGAGAACGGUGCAGGACGACUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAGGUAGUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUCUUAG ..........(((.((((.((((((.((((.(..((....(((.((((((........))))))))).....))..).)))).)))))))))).))).((((((.....))))))..... ( -47.60) >DroVir_CAF1 1352 120 - 1 AAUCGGCUUAAAGAGCAGUGCACGAGGAUUUCUCGCGGAAGGGAUUUACCUUCUGGCUGGUGAAACCAGUCAAUAGGUAAUCCUCCUGCUCGUGCUCCGAUAUAUAUUUCUUCAUUUUGU .(((((......((((((.....(((((....((.(....).)).((((((..(((((((.....)))))))..))))))))))))))))).....)))))................... ( -43.90) >DroGri_CAF1 1780 120 - 1 GAUCUGCUUAGAGAGCCGUGCACGAGGACUUCUCACGGAAGGGGUUUACCUUUUGGCUGGUGAAACCAGUCAAUAGAUAAUCCUCCUGCUCGUGCUCCGAUAUAUAUUUCUUCAUUUUAU ..........(.((((((.(((.(((((((((.....))))........((.((((((((.....)))))))).))....))))).))).)).)))))...................... ( -38.00) >DroWil_CAF1 1784 120 - 1 GACCGCCUUAAAGCACUGUGCAUGAGGAUUUCUCACGGAACGGAUUAACCUUUUGGCUGGUGAAACCAGUCAAUAGAUAAUCCUCCUGCUCAUGCUCUGUGAUAUAUUUCAUCAUUUUGU .............(((.(.(((((((((....))..(((..((((((..((.((((((((.....)))))))).)).)))))))))..))))))).).)))................... ( -35.70) >DroMoj_CAF1 1696 120 - 1 AACCUGCUUAAAGAGCAGUGCACGACGACUUCUCACGGAAUGGGUUUACCUUCUGACUGGUGAAGCCAGUCAACAGAUAAUCCUCCUGCUCGUGCUCCGAUAUAUACUUCUUCAUUUUAU ...((((((...)))))).(((((((((....))..(((..(((((...((..(((((((.....)))))))..))..)))))))).).))))))......................... ( -32.90) >DroAna_CAF1 1946 120 - 1 CACCAUCUUAAAGUACGGUGCAUGAUGACUUCUCGCGGAAGGGAUUAACCUUCUGGCUGGUGAAUCCAGUCAGCAGGUAGUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUUUUAG ........((((((((((.((((((.((....))((((..(((((((.(((.((((((((.....)))))))).)))))))))).)))))))))).))))((((.....)))))))))). ( -49.00) >consensus AACCUGCUUAAAGAACAGUGCACGAGGACUUCUCACGGAAGGGAUUUACCUUCUGGCUGGUGAAACCAGUCAACAGAUAAUCCUCCUGCUCGUGCUCCGAGAUAUACUUCAUCAUUUUAU .................(.((((((.((....))..(((..(((((...((.((((((((.....)))))))).))..))))))))...)))))).)....................... (-27.07 = -26.52 + -0.55)

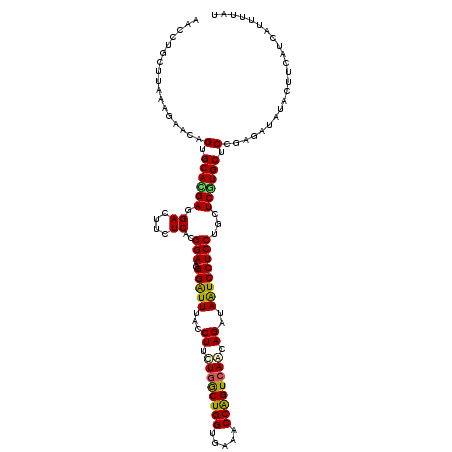
| Location | 4,414,300 – 4,414,402 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 72.14 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -13.77 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4414300 102 + 20766785 CUACCUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAGUCGUCCUGCACCGUUCUCUAAGGAGGCGGUG-AGGAUGGUCCGCCGUUGCC ......((.....(((((((((........))))))))).....(((.((..(((((((.((((((.(((....)))))))))-))))))))))))....)). ( -41.50) >DroVir_CAF1 1392 86 + 1 UUACCUAUUGACUGGUUUCACCAGCCAGAAGGUAAAUCCCUUCCGCGAGAAAUCCUCGUGCACUGCUCUUUAAGCCGAUUC--------GAAUC--------- ((((((.(((.((((.....)))).))).))))))........((((((.....))))))..(.(((.....))).)....--------.....--------- ( -23.20) >DroPse_CAF1 2555 83 + 1 UUACCUUUUGACUGGAUUCACCAGCCAGAAGGUAAAUCCAUUCCGUGAAAAGUCUUCAUGCACCGUUCUUUAAGGAGGAGUUU-------------------- ((((((((((.((((.....)))).)))))))))).(((....((((((.....))))))..((.........)).)))....-------------------- ( -25.70) >DroSim_CAF1 1825 102 + 1 UUACCUGCUUACCGGAUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAAUCGUCGUGCACAGUCCUCUAAGGAGGCGGUG-AGGAUUGUCCGCCGUUGCC ......((..((.(((((((((........))))))))).....((((....)))).(((.(((((((((...(....)...)-)))))))).))).)).)). ( -35.90) >DroMoj_CAF1 1736 94 + 1 UUAUCUGUUGACUGGCUUCACCAGUCAGAAGGUAAACCCAUUCCGUGAGAAGUCGUCGUGCACUGCUCUUUAAGCAGGUUUUUAUGGAUGGAUC--------- ((((((.((((((((.....)))))))).))))))..((((.(((((((((((......)).(((((.....))))).)))))))))))))...--------- ( -34.80) >DroPer_CAF1 2662 83 + 1 UUACCUUUUGACUGGAUUCACCAGCCAGAAGGUAAAUCCAUUCCGUGAAAAGUCUUCAUGCACCGUUCUUUAAGGAGGAGUUU-------------------- ((((((((((.((((.....)))).)))))))))).(((....((((((.....))))))..((.........)).)))....-------------------- ( -25.70) >consensus UUACCUGUUGACUGGAUUCACCAGCCAGAAGGUAAAUCCAUUCCGCGAGAAGUCGUCGUGCACCGUUCUUUAAGGAGGAGUUU______GG__C_________ ((((((.(((.((((.....)))).))).))))))........(((((.......)))))......((((....))))......................... (-13.77 = -13.97 + 0.20)

| Location | 4,414,300 – 4,414,402 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 72.14 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.17 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4414300 102 - 20766785 GGCAACGGCGGACCAUCCU-CACCGCCUCCUUAGAGAACGGUGCAGGACGACUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAGGUAG (....)((....)).((((-(((((.(((....)))..))))).))))..(((((((((....(((.((((((........))))))))).))))).)))).. ( -38.70) >DroVir_CAF1 1392 86 - 1 ---------GAUUC--------GAAUCGGCUUAAAGAGCAGUGCACGAGGAUUUCUCGCGGAAGGGAUUUACCUUCUGGCUGGUGAAACCAGUCAAUAGGUAA ---------.....--------(((((.((((...))))....(.((((.....)))).).....)))))((((..(((((((.....)))))))..)))).. ( -26.10) >DroPse_CAF1 2555 83 - 1 --------------------AAACUCCUCCUUAAAGAACGGUGCAUGAAGACUUUUCACGGAAUGGAUUUACCUUCUGGCUGGUGAAUCCAGUCAAAAGGUAA --------------------....((((((.....(((.(((........))).)))..)))..))).(((((((.(((((((.....))))))).))))))) ( -23.80) >DroSim_CAF1 1825 102 - 1 GGCAACGGCGGACAAUCCU-CACCGCCUCCUUAGAGGACUGUGCACGACGAUUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAAUCCGGUAAGCAGGUAA (....)(((((........-..)))))(((.....)))((((...((.(((....)))))....(((((((((........))))))))).....)))).... ( -34.90) >DroMoj_CAF1 1736 94 - 1 ---------GAUCCAUCCAUAAAAACCUGCUUAAAGAGCAGUGCACGACGACUUCUCACGGAAUGGGUUUACCUUCUGACUGGUGAAGCCAGUCAACAGAUAA ---------(((((((((........((((((...)))))).....((.(....)))..)).)))))))...((..(((((((.....)))))))..)).... ( -26.20) >DroPer_CAF1 2662 83 - 1 --------------------AAACUCCUCCUUAAAGAACGGUGCAUGAAGACUUUUCACGGAAUGGAUUUACCUUCUGGCUGGUGAAUCCAGUCAAAAGGUAA --------------------....((((((.....(((.(((........))).)))..)))..))).(((((((.(((((((.....))))))).))))))) ( -23.80) >consensus _________G__CC______AAACACCUCCUUAAAGAACGGUGCACGACGACUUCUCACGGAAGGGAUUUACCUUCUGGCUGGUGAAUCCAGUCAACAGGUAA .........................((...((...(((.(((........))).)))...))..))..((((((..(((((((.....)))))))..)))))) (-16.22 = -16.17 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:33 2006