| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,407,007 – 4,407,108 |
| Length | 101 |
| Max. P | 0.708589 |

| Location | 4,407,007 – 4,407,108 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
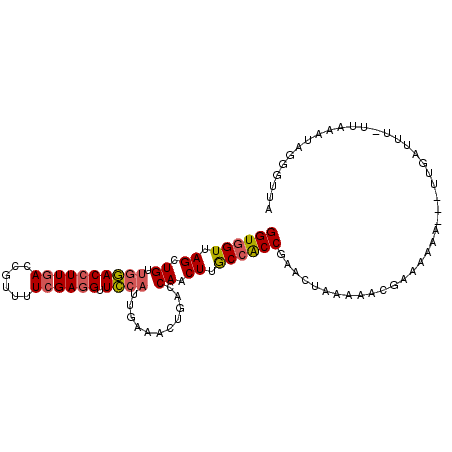
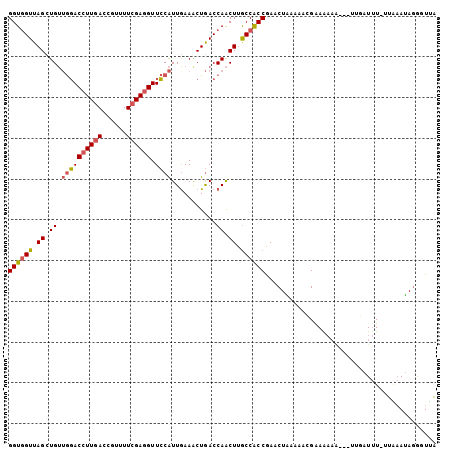
>2R_DroMel_CAF1 4407007 101 - 20766785 GGUGGUUAGCUGUUGGACCUUGACCGUUUUCGAGGUUCCAUUGAAACUGACCAACUUGCCACCGAACUAAAAACGAAGAAA---UUGAUUU-UUAAAUAGGUUUA (((((((((....((((((((((......)))))).))))......)))))).....)))(((....((((((..((....---))..)))-)))....)))... ( -25.80) >DroVir_CAF1 16431 101 - 1 GGUUGUUAGCUGGGAUACCUUCACAGUUUUCGAUGUUGCGUUGAAACUAACCAGCUUGCCGCCGAACUGUUAAUGAAAUUGUAAUUAAU-U-GGGCACA--GUUC (((.((.((((((((.....))..(((((.(((((...))))))))))..)))))).)).)))(((((((......(((((....))))-)-....)))--)))) ( -29.80) >DroSec_CAF1 16977 101 - 1 GGUGGUUAGCUGUUGGACCUUGACCGUUUUCGAGGUUCCAUUGAAGCUGACCAACUUGCCACCGAACUAAAAACGAAAAAA---AUUAUUU-UUAAAUAGGGUUA (((((((((((..((((((((((......)))))).))))....)))))))).....))).((....((((((..(.....---.)..)))-)))....)).... ( -30.60) >DroSim_CAF1 16847 101 - 1 GGUGGUUAGCUGUUGGACCUUGACCGUUUUCGAGGUUCCAUUGAAGCUGACCAACUUGCCACCGAACUAAAAACGAAAAAA---UGGAUUU-UUAAAUAGGGUUA (((((((((((..((((((((((......)))))).))))....)))))))).....))).((....(((((((.(.....---).).)))-)))....)).... ( -29.50) >DroEre_CAF1 17081 101 - 1 GGUGGUUAGCUGUUGGACCUUGAUGGUUUUCGAGGUUCCAUUGAAACUGACCAACUUACCACCAAACUGAAAUCGGAAAAU---GUGAUUU-UUAAAUAGAUUUA ((((((.....((((((((((((......)))))))((....))......)))))..))))))......(((((.((((((---...))))-)).....))))). ( -26.10) >DroYak_CAF1 17013 102 - 1 GGUGGUUAGCUGUUGGACCUUGACCGUUUUCGAGGUUCCAUUAAAACUGACCAACUUGCCACCAAACUAAAAACGGAAAAG---GUGAUUUGAUUAAUAGGUUAA ..(((((((....((((((((((......)))))).))))......)))))))......((((.................)---))).................. ( -23.93) >consensus GGUGGUUAGCUGUUGGACCUUGACCGUUUUCGAGGUUCCAUUGAAACUGACCAACUUGCCACCGAACUAAAAACGAAAAAA___UUGAUUU_UUAAAUAGGGUUA ((((((.((.((.((((((((((......)))))).))))...........)).)).)))))).......................................... (-18.44 = -18.72 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:27 2006