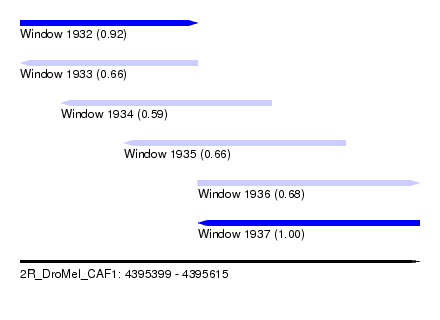
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,395,399 – 4,395,615 |
| Length | 216 |
| Max. P | 0.999826 |

| Location | 4,395,399 – 4,395,495 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4395399 96 + 20766785 AGAACCUUAAAAACC-AUUCACUAAUACCUC-CACAGGAACAAGAGAUGGAGAAGCAGAAGCUGCUCUUCCAUCAGGCACGUCUCUCCAAUCGUGGCG ...............-...........(((.-...))).......((((((..(((((...)))))..)))))).(.((((..........)))).). ( -23.50) >DroVir_CAF1 5259 87 + 1 ----------AAUCAAAUGCUCUAAAUUAUCGU-UAGGAACAAGAAAUGGAGAAACAGAAGUUGCUCUUCCAUCAAGCGCGUCUUUCGAAUCGUGGCG ----------........(((........(((.-.(((........((((((.(((....)))....))))))........)))..))).....))). ( -11.21) >DroSec_CAF1 5408 86 + 1 ----------AAACA-AUUCACUAAUACCUC-CACAGGAACAAGAGAUGGAGAAGCAGAAGCUGCUCUUCCAUCAGGCACGUCUCUCCAAUCGUGGCG ----------.....-...........(((.-...))).......((((((..(((((...)))))..)))))).(.((((..........)))).). ( -23.50) >DroSim_CAF1 5309 86 + 1 ----------AAACC-AUUCACUAAUUCCUC-CACAGGAACAAGAGAUGGAGAAGCAGAAGCUGCUCUUCCAUCAGGCACGUCUCUCCAAUCGUGGCG ----------.....-.....((..(((((.-...)))))..)).((((((..(((((...)))))..)))))).(.((((..........)))).). ( -26.40) >DroYak_CAF1 5416 96 + 1 UUUGCCUUAAAAACC-AUUCAUUAAUACCUC-UACAGGAACAAGAGAUGGAGAAGCAGAAACUGCUCUUCCAUCAGGCACGUCUCUCCAAUCGUGGCG ..(((((........-...........(((.-...))).......((((((..(((((...)))))..)))))))))))((((.(.......).)))) ( -26.90) >DroPer_CAF1 5081 78 + 1 ----------UAACC---------AUCAUUC-GAUAGGAACAAGAGAUGGAAAAGCAGAAGCUGCUCUUCCAUCAGGCCCGCCUCUCGAAUCGUGGCG ----------...((---------((.((((-((.(((.......(((((((.(((((...))))).))))))).......))).)))))).)))).. ( -31.64) >consensus __________AAACC_AUUCACUAAUACCUC_CACAGGAACAAGAGAUGGAGAAGCAGAAGCUGCUCUUCCAUCAGGCACGUCUCUCCAAUCGUGGCG .............................................(((((((.(((((...))))).))))))).(.((((..........)))).). (-17.64 = -17.92 + 0.28)


| Location | 4,395,399 – 4,395,495 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.43 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4395399 96 - 20766785 CGCCACGAUUGGAGAGACGUGCCUGAUGGAAGAGCAGCUUCUGCUUCUCCAUCUCUUGUUCCUGUG-GAGGUAUUAGUGAAU-GGUUUUUAAGGUUCU .(((.(.((.(((((((.......((((((..(((((...)))))..))))))))))..))).)).-).)))......((((-..(....)..)))). ( -25.51) >DroVir_CAF1 5259 87 - 1 CGCCACGAUUCGAAAGACGCGCUUGAUGGAAGAGCAACUUCUGUUUCUCCAUUUCUUGUUCCUA-ACGAUAAUUUAGAGCAUUUGAUU---------- ......(..((....))..)((((((((((..((((.....))))..))))))..((((.....-)))).......))))........---------- ( -17.40) >DroSec_CAF1 5408 86 - 1 CGCCACGAUUGGAGAGACGUGCCUGAUGGAAGAGCAGCUUCUGCUUCUCCAUCUCUUGUUCCUGUG-GAGGUAUUAGUGAAU-UGUUU---------- .(((.(.((.(((((((.......((((((..(((((...)))))..))))))))))..))).)).-).)))..........-.....---------- ( -25.31) >DroSim_CAF1 5309 86 - 1 CGCCACGAUUGGAGAGACGUGCCUGAUGGAAGAGCAGCUUCUGCUUCUCCAUCUCUUGUUCCUGUG-GAGGAAUUAGUGAAU-GGUUU---------- ..(((....)))........(((.((((((..(((((...)))))..)))))).((.((((((...-.)))))).)).....-)))..---------- ( -26.50) >DroYak_CAF1 5416 96 - 1 CGCCACGAUUGGAGAGACGUGCCUGAUGGAAGAGCAGUUUCUGCUUCUCCAUCUCUUGUUCCUGUA-GAGGUAUUAAUGAAU-GGUUUUUAAGGCAAA .(((((..(((.((..(((.....((((((..(((((...)))))..))))))...)))..)).))-)..)).....((((.-....)))).)))... ( -27.80) >DroPer_CAF1 5081 78 - 1 CGCCACGAUUCGAGAGGCGGGCCUGAUGGAAGAGCAGCUUCUGCUUUUCCAUCUCUUGUUCCUAUC-GAAUGAU---------GGUUA---------- .((((..((((((.(((((((...(((((((((((((...))))))))))))).))))..))).))-))))..)---------)))..---------- ( -36.10) >consensus CGCCACGAUUGGAGAGACGUGCCUGAUGGAAGAGCAGCUUCUGCUUCUCCAUCUCUUGUUCCUGUG_GAGGUAUUAGUGAAU_GGUUU__________ ............((..(((.....((((((..(((((...)))))..))))))...)))..))................................... (-17.51 = -17.43 + -0.08)

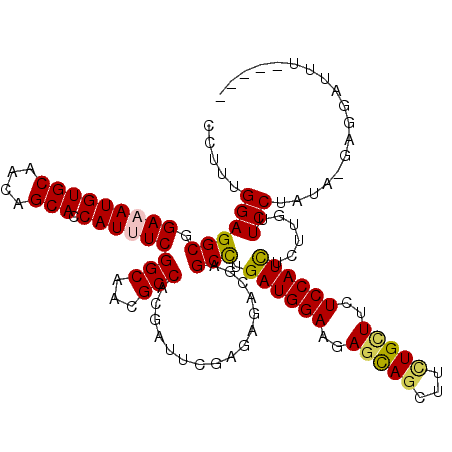
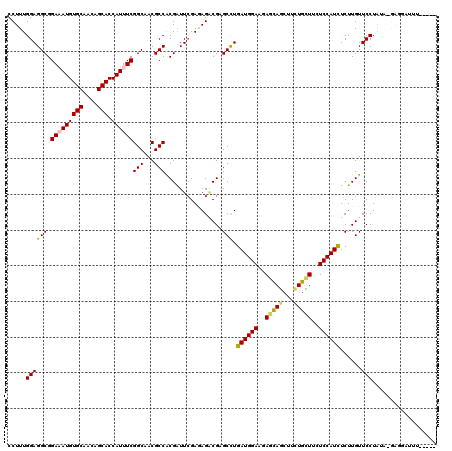
| Location | 4,395,421 – 4,395,535 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.33 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -28.80 |
| Energy contribution | -28.50 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4395421 114 - 20766785 CCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUGGAGAGACGUGCCUGAUGGAAGAGCAGCUUCUGCUUCUCCAUCUCUUGUUCCUGUG-GAGGUAUU----- (((..(.(((..(((((((((....))).))))))((((.(.(((....)))...)...)))).((((((..(((((...)))))..)))))).......))).).-.)))....----- ( -42.00) >DroVir_CAF1 5272 114 - 1 CCUUUGGAGGCGGAUAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGAAAGACGCGCUUGAUGGAAGAGCAACUUCUGUUUCUCCAUUUCUUGUUCCUA-ACGAUAAUUU----- .....(((((((((...((((....))))...)))(....)))).((..((....))..))...((((((..((((.....))))..))))))......)))..-..........----- ( -30.30) >DroGri_CAF1 6155 120 - 1 CCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGAGAGUCGAGCCUGAUGGAACAGCAGCUUCUGUUUCUCCAUUUCUUGUUCCUGUGCGAUCAUGUCAAAA ..(((((..((.(((((((((....))).)))))).))..(((.((((((....))))((((..((((((..(((((...)))))..))))))....))))..)))))......))))). ( -35.20) >DroWil_CAF1 6182 111 - 1 CCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCGCGGUUCGAAAGACGAGCCUGAUGGAAUAAUAGCUUUUGCUUCUCCAUCUCUUGUUCCUACA-AGGGG-------- ((((((..(((.(((((((((....))).))))))(....))))..((((((.....)))))).((((((.....(((....)))..))))))...........))-)))).-------- ( -41.50) >DroMoj_CAF1 5325 107 - 1 CCUUUGGAGGCGGAUAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGACAGACGCGCUUGAUGGAACAGUAGCUUCUGUUUCUCCAUUUCUUGUUCCUG--------UUU----- ...(((..((((((...((((....))))...)))(....)))).)))...(((((..(((...((((((((((......))))...))))))...)))..)))--------)).----- ( -29.50) >DroPer_CAF1 5086 113 - 1 CCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGAGAGGCGGGCCUGAUGGAAGAGCAGCUUCUGCUUUUCCAUCUCUUGUUCCUAUC-GAAUGAU------ ....(((..((.(((((((((....))).)))))).))....)))..((((((.(((((((...(((((((((((((...))))))))))))).))))..))).))-))))...------ ( -47.60) >consensus CCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGAGAGACGAGCCUGAUGGAAGAGCAGCUUCUGCUUCUCCAUCUCUUGUUCCUAUA_GAGGAUUU_____ .....((((((.(((((((((....))).))))))(((...)))................))).((((((..(((((...)))))..))))))......))).................. (-28.80 = -28.50 + -0.30)

| Location | 4,395,455 – 4,395,575 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -34.67 |
| Energy contribution | -34.12 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4395455 120 - 20766785 GCCGAGAUUGAGUGUCGUCAUUACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUGGAGAGACGUGCCUGAUGGAAGAGCAGCUU ((((((((.(.((((.((((((.((..(((((((((....)))))))))..)).))))....)).)))))))))))))....(((....))).(((.(.(((((......)).))))))) ( -43.30) >DroVir_CAF1 5306 120 - 1 GCCCAAAUUGAGUGUCGUCAUGACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAUAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGAAAGACGCGCUUGAUGGAAGAGCAACUU (((...((..((((.((((.......((((((((((....))))))))))(((((..((((....))))......(((...)))...)))))...))))))))..))....).))..... ( -42.10) >DroGri_CAF1 6195 120 - 1 GCCCAAAUUGAGUGUCGUCAUGACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGAGAGUCGAGCCUGAUGGAACAGCAGCUU ((.........(((....)))..(((((((((((((....))))))))(((.(((((((((....))).))))))(((...))).(((((....))))).))).)))))....))..... ( -40.80) >DroWil_CAF1 6213 120 - 1 GCCGAGAUUGAGUGUUGUCAUUACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCGCGGUUCGAAAGACGAGCCUGAUGGAAUAAUAGCUU ((((((((.(.(((((((((((.((..(((((((((....)))))))))..)).))))....))))))))))))))))....((((((((((.....)))))).).)))........... ( -51.60) >DroMoj_CAF1 5352 120 - 1 GCCCAAAUUGAGUGUCGUCAUGACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAUAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGACAGACGCGCUUGAUGGAACAGUAGCUU ..(((..(..((((.((((.((....((((((((((....))))))))))(((((..((((....))))......(((...)))...))))).))))))))))..))))........... ( -41.80) >DroAna_CAF1 6608 120 - 1 GCCGAGAUUGAGUGUCGUCAUUACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACACCACGAUUGGAGAGACGUGCCUGAUGGAAGAGCAGUUU ((((((((.(.((((.((((((.((..(((((((((....)))))))))..)).))))....)).)))))))))))))....(((....)))..((((.(((((......)).))))))) ( -44.80) >consensus GCCCAAAUUGAGUGUCGUCAUGACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAACGCCACGAUUCGAGAGACGAGCCUGAUGGAAGAGCAGCUU .....(((((.((((.(((.....(..(((((((((....)))))))))..)(((((((((....))).))))))))).))))..)))))...(((..((.((....))....))..))) (-34.67 = -34.12 + -0.55)



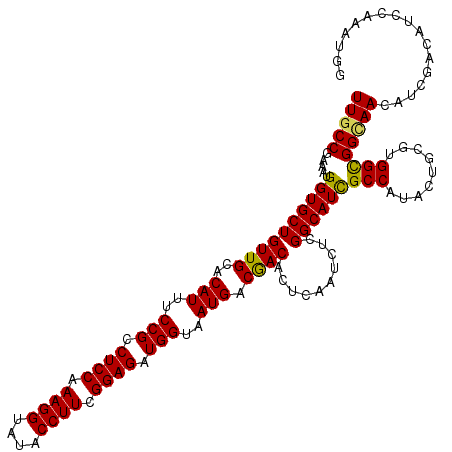
| Location | 4,395,495 – 4,395,615 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -33.18 |
| Energy contribution | -32.77 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4395495 120 + 20766785 UUGCCGAAAUGGUGCUGUUGCACAUUUCCGCCUCCAAAGGUAUACCUUCGGAGAUGGUAAUGACGACACUCAAUCUCGGCAUCGCCAUUUUGCGCGGCGGCAACAUCGACAUCCAAAUGG ((((((.....((((....))))....((((((((.((((....)))).))))((((..(((.(((.........))).)))..)))).....))))))))))................. ( -40.80) >DroVir_CAF1 5346 120 + 1 UUGCCGAAAUGGUGCUGUUGCACAUAUCCGCCUCCAAAGGUAUACCUUCGGAGAUGGUCAUGACGACACUCAAUUUGGGCAUCGCCAUAUUGCGUGGCGGAAACAUUGAUAUACAGAUGG .((((.((((.(.(.(((((..(((..(((.((((.((((....)))).)))).)))..))).)))))).).)))).))))(((((((.....)))))))...((((........)))). ( -41.40) >DroGri_CAF1 6235 120 + 1 UUGCCGAAAUGGUGCUGUUGCACAUUUCCGCCUCCAAAGGUAUACCUUCGGAGAUGGUCAUGACGACACUCAAUUUGGGCAUCGCCAUAUUGCGGGGUGGCAACAUCGACAUCCAAAUGG .((((.((((.(.(.(((((..(((..(((.((((.((((....)))).)))).)))..))).)))))).).)))).))))((((......))))((((....))))............. ( -39.80) >DroWil_CAF1 6253 120 + 1 UUGCCGAAAUGGUGCUGUUGCACAUUUCCGCCUCCAAAGGUAUACCUUCGGAGAUGGUAAUGACAACACUCAAUCUCGGCAUUGCCAUUCUACGUGGCGGUAACAUUGACAUCCAAAUGG ..(((((.((.(.(.(((((..((((.(((.((((.((((....)))).)))).))).)))).)))))).).)).)))))((((((((.....))))))))..((((........)))). ( -41.50) >DroMoj_CAF1 5392 120 + 1 UUGCCGAAAUGGUGCUGUUGCACAUAUCCGCCUCCAAAGGUAUACCUUCGGAGAUGGUCAUGACGACACUCAAUUUGGGCAUCGCCAUACUGCGUGGCGGUAACAUCGACAUUCAAAUGG ..(((.((((.(.(.(((((..(((..(((.((((.((((....)))).)))).)))..))).)))))).).)))).)))((((((((.....))))))))................... ( -40.50) >DroAna_CAF1 6648 120 + 1 UUGCCGAAAUGGUGCUGUUGCACAUUUCCGCCUCCAAAGGUAUACCUUCGGAGAUGGUAAUGACGACACUCAAUCUCGGCAUCGCCAUCCUGAGAGGCGGCAACAUCGACAUCCAAAUGG (((((((((((.(((....))))))))).(((((...(((....)))((((.(((((..(((.(((.........))).)))..)))))))))))))))))))................. ( -44.80) >consensus UUGCCGAAAUGGUGCUGUUGCACAUUUCCGCCUCCAAAGGUAUACCUUCGGAGAUGGUAAUGACGACACUCAAUCUCGGCAUCGCCAUACUGCGUGGCGGCAACAUCGACAUCCAAAUGG (((((.....((((((((((..(((..(((.((((.((((....)))).)))).)))..))).))))..........))))))(((.........))))))))................. (-33.18 = -32.77 + -0.42)


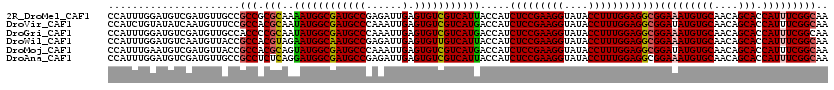
| Location | 4,395,495 – 4,395,615 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -48.08 |
| Consensus MFE | -43.17 |
| Energy contribution | -42.87 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.18 |
| SVM RNA-class probability | 0.999826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4395495 120 - 20766785 CCAUUUGGAUGUCGAUGUUGCCGCCGCGCAAAAUGGCGAUGCCGAGAUUGAGUGUCGUCAUUACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAA ((....)).((((((((.(((.(..(((((.(((((((((((((....)).))))))))))).((..(((((((((....)))))))))..))...)))))..).))).))..)))))). ( -49.50) >DroVir_CAF1 5346 120 - 1 CCAUCUGUAUAUCAAUGUUUCCGCCACGCAAUAUGGCGAUGCCCAAAUUGAGUGUCGUCAUGACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAUAUGUGCAACAGCACCAUUUCGGCAA ......................(((.(((..(((((((((((.(.....).))))))))))).....(((((((((....))))))))))))((.((((((....))).))).))))).. ( -44.00) >DroGri_CAF1 6235 120 - 1 CCAUUUGGAUGUCGAUGUUGCCACCCCGCAAUAUGGCGAUGCCCAAAUUGAGUGUCGUCAUGACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAA ((....)).((((((((((((((..((((..(((((((((((.(.....).))))))))))).....(((((((((....)))))))))))))...)).))))))........)))))). ( -49.50) >DroWil_CAF1 6253 120 - 1 CCAUUUGGAUGUCAAUGUUACCGCCACGUAGAAUGGCAAUGCCGAGAUUGAGUGUUGUCAUUACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAA ((....)).(((((...((((......))))..))))).(((((((((.(.(((((((((((.((..(((((((((....)))))))))..)).))))....))))))))))))))))). ( -49.00) >DroMoj_CAF1 5392 120 - 1 CCAUUUGAAUGUCGAUGUUACCGCCACGCAGUAUGGCGAUGCCCAAAUUGAGUGUCGUCAUGACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAUAUGUGCAACAGCACCAUUUCGGCAA .........((((((.....(((((......(((((((((((.(.....).)))))))))))......((((((((....)))))))))))))....((((....))))....)))))). ( -47.30) >DroAna_CAF1 6648 120 - 1 CCAUUUGGAUGUCGAUGUUGCCGCCUCUCAGGAUGGCGAUGCCGAGAUUGAGUGUCGUCAUUACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAA ((....)).((((((.(((.(((((((...((((((..(((.(((.((...)).))).)))..)))))).((((((....))))))))))))).)))((((....))))....)))))). ( -49.20) >consensus CCAUUUGGAUGUCGAUGUUGCCGCCACGCAAUAUGGCGAUGCCCAAAUUGAGUGUCGUCAUGACCAUCUCCGAAGGUAUACCUUUGGAGGCGGAAAUGUGCAACAGCACCAUUUCGGCAA ......................(((.(((..((((((((((((......).))))))))))).....(((((((((....))))))))))))(((((((((....))).))))))))).. (-43.17 = -42.87 + -0.30)
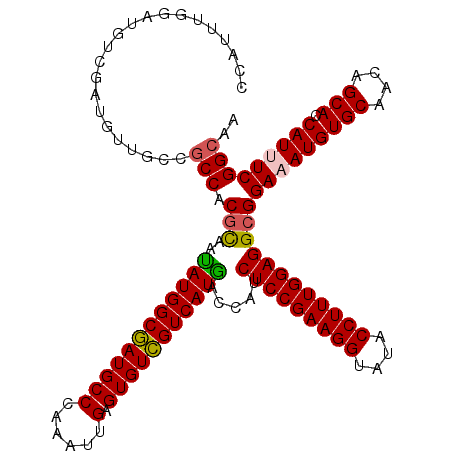
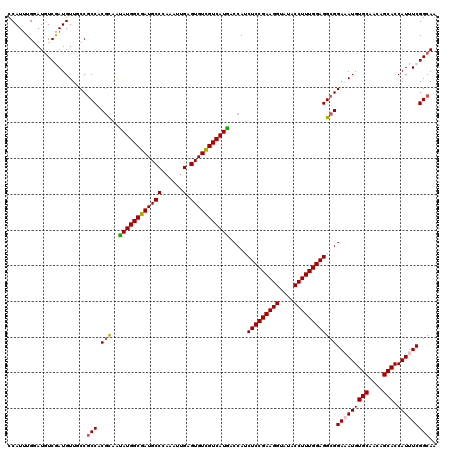
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:23 2006