| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,394,116 – 4,394,263 |
| Length | 147 |
| Max. P | 0.953633 |

| Location | 4,394,116 – 4,394,236 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.11 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -28.75 |
| Energy contribution | -28.20 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4394116 120 + 20766785 AUCGACCAUCCACAACAGCAGAGCAAGAAUGUCUACAGGAGCGUUGUGUCGAUCCAAAGGAAGCGUGCCGUAAUCGCAUGCUUCCGUCAGACAUCGCUGCAUUCUCUACCCAGGUUAUUA ...((((..........((((.....((.(((((((.(((.((......)).)))...((((((((((.......)))))))))))).))))))).))))............)))).... ( -36.65) >DroVir_CAF1 3668 120 + 1 AUCGACCAUCCACAACAGCAGAGCAAAAAUGUCUACAGGAGCGUUGUAUCGAUCCAAAGGAAACGUGCCGUCAUUGCAUGUUUCCGUCAGACAUCGCUACAUUCUCUACCCAGGUUAUUA ...((((..............(((....((((((((.(((.((......)).)))...((((((((((.......)))))))))))).)))))).)))..............)))).... ( -32.39) >DroPse_CAF1 3688 120 + 1 AUCGACCAUCCGCAACAGCAGAGCAAGAAUGUGUACAGGAGCGUUGUGUCGAUCCAAAGGAAGCGUGCCGUAAUCGCAUGUUUCCGUCAGACAUCGCUACACUCUCUACCCAGGUUAAUG ...((((.((.((....)).))...(((..((((.....((((..((((((((.....((((((((((.......))))))))))))).)))))))))))))..))).....)))).... ( -35.60) >DroWil_CAF1 3744 120 + 1 AUCGACCAUCCACAACAGCAGAGCAAAAAUGUCUACAGGAGCGUUGUAUCGAUACAAAGGAAACGUGCAGUUAUCGCAUGUUUCCGCCAAACAUCGUUACAUUCUCUACCCAGGUUAUUA ...((((..........(.(((.((....))))).).((((...((((.((((.....((((((((((.......)))))))))).......)))).))))..)))).....)))).... ( -30.70) >DroMoj_CAF1 3702 120 + 1 AUCGACCAUCCACAACAGCAGAGCAAAAAUGUCUACAGGAGCGUUGUGUCAAUACAAAGGAAGCGUGCCGUUAUCGCAUGUUUCCGUCAGACAUCGCUACAUUCUCUACCCAGGUUAUUA ...((((..............(((....((((((((.......(((((....))))).((((((((((.......)))))))))))).)))))).)))..............)))).... ( -28.89) >DroPer_CAF1 3690 120 + 1 AUCGACCAUCCGCAACAGCAGAGCAAGAAUGUGUACAGGAGCGUUGUGUCGAUCCAAAGGAAGCGUGCCGUAAUCGCAUGUUUCCGUCAGACAUCGCUACACUCUCUACCCAGGUUAAUG ...((((.((.((....)).))...(((..((((.....((((..((((((((.....((((((((((.......))))))))))))).)))))))))))))..))).....)))).... ( -35.60) >consensus AUCGACCAUCCACAACAGCAGAGCAAAAAUGUCUACAGGAGCGUUGUGUCGAUCCAAAGGAAGCGUGCCGUAAUCGCAUGUUUCCGUCAGACAUCGCUACAUUCUCUACCCAGGUUAUUA ...((((..........((...)).............((((...((((.((((.....((((((((((.......)))))))))).......)))).))))..)))).....)))).... (-28.75 = -28.20 + -0.55)

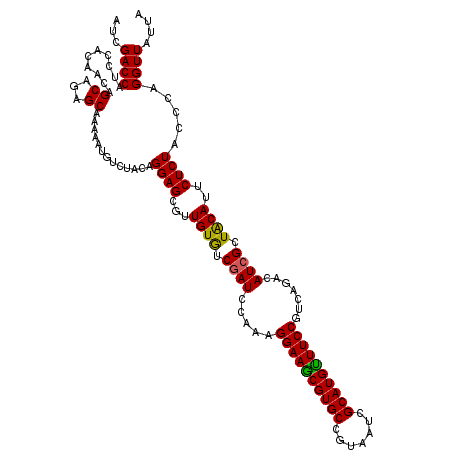

| Location | 4,394,156 – 4,394,263 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.13 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953633 |
| Prediction | RNA |
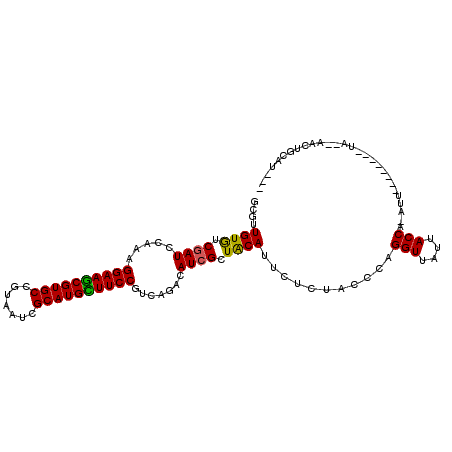
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4394156 107 + 20766785 GCGUUGUGUCGAUCCAAAGGAAGCGUGCCGUAAUCGCAUGCUUCCGUCAGACAUCGCUGCAUUCUCUACCCAGGUUAUUACCG-AUUGCAGUUAACA--GAUUGCAUCUU ((((.((((((((.....((((((((((.......))))))))))))).))))).)).))............(((....)))(-((.((((((....--))))))))).. ( -35.50) >DroVir_CAF1 3708 99 + 1 GCGUUGUAUCGAUCCAAAGGAAACGUGCCGUCAUUGCAUGUUUCCGUCAGACAUCGCUACAUUCUCUACCCAGGUUAUUACCA-ACU--------UAUUAACUACUGC-- ((..((((.((((.....((((((((((.......)))))))))).......)))).))))...........(((....))).-...--------...........))-- ( -23.40) >DroGri_CAF1 3967 105 + 1 GUGUUGUGUCGAUCCAAAGGAAGCGUGCCGUAAUCGCAUGCUUCCGUCAGACAUCGUUACAUUCUCUGCCCAGGUUAUUACCACAAU-----UAAUAUCAACUGCAUUUU (((((((((((((.....((((((((((.......))))))))))))).))))...................(((....))).....-----......)))).))..... ( -26.00) >DroWil_CAF1 3784 99 + 1 GCGUUGUAUCGAUACAAAGGAAACGUGCAGUUAUCGCAUGUUUCCGCCAAACAUCGUUACAUUCUCUACCCAGGUUAUUACCA-ACAGAA----AUA--GAUUACU---- ..((((((.((((.....((((((((((.......)))))))))).......)))).)))............(((....))))-))....----...--.......---- ( -24.00) >DroMoj_CAF1 3742 99 + 1 GCGUUGUGUCAAUACAAAGGAAGCGUGCCGUUAUCGCAUGUUUCCGUCAGACAUCGCUACAUUCUCUACCCAGGUUAUUACCG-AUA--------UAUUCACUGCAUU-- ((((.(((((........((((((((((.......))))))))))....))))).))...............(((....))).-...--------........))...-- ( -24.60) >DroAna_CAF1 5039 85 + 1 GCGUUGUGUCGAUCCAAAGGAAGCGUGCCGUAAUCGCAUGCUUCCGUCAGACAUCGCUACAUUCUCUACCCAGGUUAUUACCG-AU------------------------ (((..((((((((.....((((((((((.......))))))))))))).))))))))...............(((....))).-..------------------------ ( -27.70) >consensus GCGUUGUGUCGAUCCAAAGGAAGCGUGCCGUAAUCGCAUGCUUCCGUCAGACAUCGCUACAUUCUCUACCCAGGUUAUUACCA_AUU________UA__AACUGCAU___ ....((((.((((.....((((((((((.......)))))))))).......)))).))))...........(((....)))............................ (-23.80 = -23.13 + -0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:17 2006