
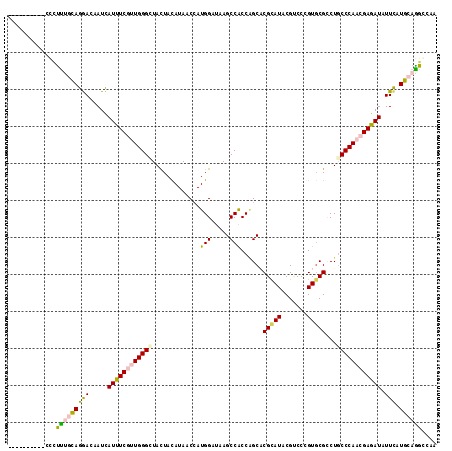
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,381,646 – 4,381,752 |
| Length | 106 |
| Max. P | 0.860416 |

| Location | 4,381,646 – 4,381,752 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -23.43 |
| Energy contribution | -23.63 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
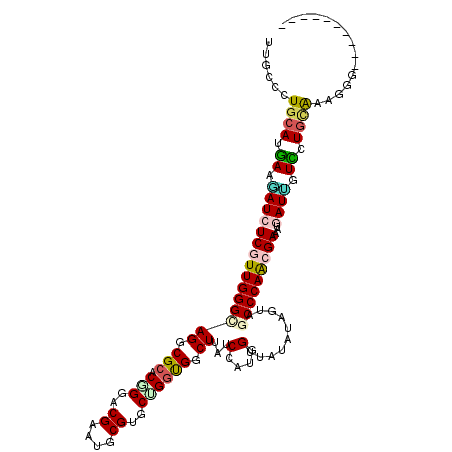
>2R_DroMel_CAF1 4381646 106 + 20766785 ----------ACCUCUAUAGGACGAUUAUCUCGCUGGGAUACUACAUAACCAUGGACAAGCCACCAGCACGCAUUCGCCCAGUGCGCCUGCCCAACGAGAUCUUCAUGCAGGGCAA ----------.(((....)))..((..((((((.((((..............(((.....))).(((..((((((.....)))))).))))))).))))))..)).(((...))). ( -28.00) >DroVir_CAF1 61924 110 + 1 ------CUCCCGCUUUGCAGAACCAUUAUUUCGUUGGGCUACUAUAUAACCAUGGAUAAGCCGCCGGCACGCAUACGUCCCGUCCGCCUGCCCAACGAGAUAUUCAUGCAAGCCAA ------.....((((.((((((....(((((((((((((..((((......))))....((.(.(((.(((....))).))).).))..)))))))))))))))).)))))))... ( -38.20) >DroGri_CAF1 8945 106 + 1 ----------CUCUUUGCAGGACAAUCAUUUCGUUGGGUUAUUAUAUAACGAUGGAUAAGCCACCGGCACGUAUACGCCCCGUGCGGCUGCCCAACGAGAUAUUCAUGCAGGCCAA ----------...((((((.((.....((((((((((((..((((......))))...((((.((((..((....))..))).).))))))))))))))))..)).)))))).... ( -33.30) >DroMoj_CAF1 4089 107 + 1 ---------UCCAUUUCCAGAACUAUCAUCUCUUUGGGCUACUAUAUAACCAUGGAUAAGCCACCAGCACGCAUACGACCCGUGCGCCUGCCCAACGAGAUAUUUAUGCAGGCCAA ---------.......((.((....))(((((.((((((.............(((........)))(((((.........)))))....)))))).))))).........)).... ( -24.90) >DroAna_CAF1 8380 106 + 1 ----------CCUUUCGCAGGACAAUCAUUUCCCUGGGCUACUACAUAACCAUGGACAAGCCGCCAGCCCGCAUUCGUCCAGUGCGCCUACCCAACGAGAUCUUCAUGCAGGGCAA ----------.......((((...........))))((((....(((....)))....))))....((((((((((((...(((....)))...)))).......)))).)))).. ( -24.01) >DroPer_CAF1 23483 116 + 1 GCUUUAUUUCCCCCCCGCAGGACAAUCAUUUCGUUGGGCUACUACAUAACCAUGGAUAAGCCACCCGCCCGCAUUCGUCCGGUGCGUCUGCCCAACGAGAUCUUCAUGCAGGGCAA ............(((.(((.((.....((((((((((((.............(((.....)))..((((((........))).)))...))))))))))))..)).))).)))... ( -36.60) >consensus __________CCCUUUGCAGGACAAUCAUUUCGUUGGGCUACUACAUAACCAUGGAUAAGCCACCAGCACGCAUACGUCCCGUGCGCCUGCCCAACGAGAUAUUCAUGCAGGCCAA .............(((((((((.....((((((((((((.............(((.....)))......(((((.......)))))...)))))))))))).))).)))))).... (-23.43 = -23.63 + 0.20)


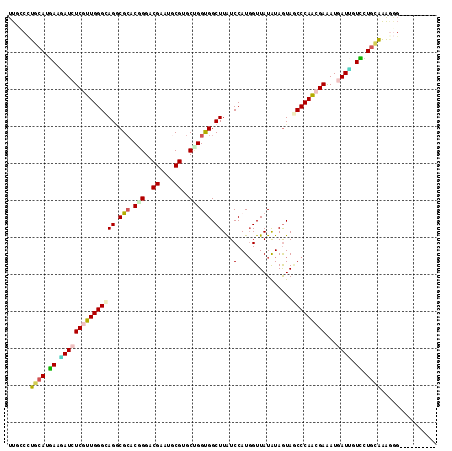
| Location | 4,381,646 – 4,381,752 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4381646 106 - 20766785 UUGCCCUGCAUGAAGAUCUCGUUGGGCAGGCGCACUGGGCGAAUGCGUGCUGGUGGCUUGUCCAUGGUUAUGUAGUAUCCCAGCGAGAUAAUCGUCCUAUAGAGGU---------- ..((((((.((((..((((((((((((((((.(((..((((....))).)..))))))))))))(((............))))))))))..))))....))).)))---------- ( -39.60) >DroVir_CAF1 61924 110 - 1 UUGGCUUGCAUGAAUAUCUCGUUGGGCAGGCGGACGGGACGUAUGCGUGCCGGCGGCUUAUCCAUGGUUAUAUAGUAGCCCAACGAAAUAAUGGUUCUGCAAAGCGGGAG------ ((.(((((((.((((...(((((((((((((.(.(((.(((....))).))).).))))..(....)..........))))))))).......))))))).)))).))..------ ( -41.00) >DroGri_CAF1 8945 106 - 1 UUGGCCUGCAUGAAUAUCUCGUUGGGCAGCCGCACGGGGCGUAUACGUGCCGGUGGCUUAUCCAUCGUUAUAUAAUAACCCAACGAAAUGAUUGUCCUGCAAAGAG---------- ......((((.((..((((((((((..((((((.(((.(((....))).)))))))))........(((((...))))))))))))...)))..)).)))).....---------- ( -37.30) >DroMoj_CAF1 4089 107 - 1 UUGGCCUGCAUAAAUAUCUCGUUGGGCAGGCGCACGGGUCGUAUGCGUGCUGGUGGCUUAUCCAUGGUUAUAUAGUAGCCCAAAGAGAUGAUAGUUCUGGAAAUGGA--------- ....((........((((((.((((((..(((((((.((...)).)))))..((((((.......))))))...)).)))))).))))))........)).......--------- ( -30.69) >DroAna_CAF1 8380 106 - 1 UUGCCCUGCAUGAAGAUCUCGUUGGGUAGGCGCACUGGACGAAUGCGGGCUGGCGGCUUGUCCAUGGUUAUGUAGUAGCCCAGGGAAAUGAUUGUCCUGCGAAAGG---------- ((((((...((((.....)))).)))))).((((..((((((...(((((.....)))))(((..((((((...))))))...))).....)))))))))).....---------- ( -33.20) >DroPer_CAF1 23483 116 - 1 UUGCCCUGCAUGAAGAUCUCGUUGGGCAGACGCACCGGACGAAUGCGGGCGGGUGGCUUAUCCAUGGUUAUGUAGUAGCCCAACGAAAUGAUUGUCCUGCGGGGGGGAAAUAAAGC ((.(((((((.((.(((((((((((((...(((.(((........)))))).((((((.......))))))......)))))))))...)))).)).))))))).))......... ( -47.00) >consensus UUGCCCUGCAUGAAGAUCUCGUUGGGCAGGCGCACGGGACGAAUGCGUGCUGGUGGCUUAUCCAUGGUUAUAUAGUAGCCCAACGAAAUGAUUGUCCUGCAAAGGG__________ ......((((.((.(((((((((((((((.(((.(((..((....))..)))))).))...(....)..........)))))))))...)))).)).))))............... (-24.02 = -24.17 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:12 2006