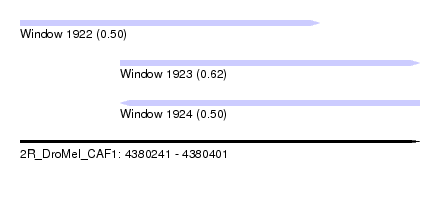
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,380,241 – 4,380,401 |
| Length | 160 |
| Max. P | 0.622340 |

| Location | 4,380,241 – 4,380,361 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -31.24 |
| Energy contribution | -29.33 |
| Covariance contribution | -1.91 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
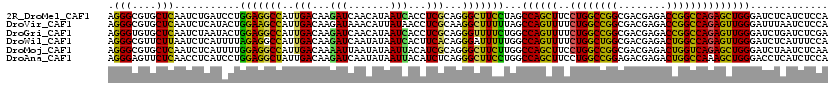
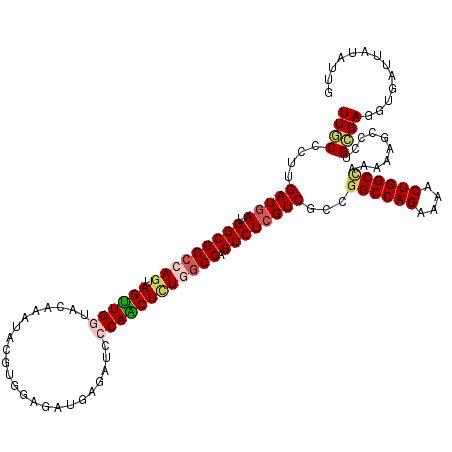
>2R_DroMel_CAF1 4380241 120 + 20766785 AGGGCGUGCUCAAUCUGAUCCUGGAGGCCAUUGACAAGAUCAACAUAAUCACCUCGCAGGGCUUCCUAGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUCUCCA .(((.(((........(((((.(((((((..(((...(((.......)))...)))...)))))))....(((((..(((((((((.....).)))))))))))))))))).))).))). ( -47.00) >DroVir_CAF1 60515 120 + 1 AGGGCGUGCUCAAUCUCAUACUGGAAGCCAUUGACAAGAUAAACAUUAUAACCUCGCAAGGCUUUUUAGCCAGUUUUCUGGCCGGCGACGAGACCGGCCAGAGUUGGGAUUUAAUCUCCA .(((.((....(((((((.((((((((((.(((.(.((..............)).))))))))).....))))).(((((((((((.....).)))))))))).)))))))..)).))). ( -39.54) >DroGri_CAF1 7358 120 + 1 AGGGUGUGCUCAAUCUAAUACUGGAGGCCAUUGACAAGAUCAACAUAAUCACCUCGCAGGGUUUUCUGGCCAGUUUUCUGGCCGGCGACGAGACCGGCCAGAGUUGGGAUCUGAUCUCGA .(((....)))..((((....))))(((((..(((..(((.......))).((.....)))))...)))))....(((((((((((.....).))))))))))(((((((...))))))) ( -38.30) >DroWil_CAF1 8121 120 + 1 AGGGCGUUCUUAAUCUCAUUUUAGAGGCCAUUGACAAGAUCAAUAUAAUCACUUCACAGGGAUUUUUGGCCAGUUUUCUGGCUGGCGACGAGACUGGCCAGAGUUGGGAUCUCAUUUCCA ..(((.((((.((......)).)))))))(((((.....)))))..............(((((((((((((((((((.(.(....).).))))))))))))).....))))))....... ( -33.10) >DroMoj_CAF1 2679 120 + 1 AGGGCGUGCUCAAUCUCAUUUUGGAGGCCAUUGACAAAAUUAAUAUAAUUACAUCGCAGGGCUUCUUGGCCAGCUUCCUGGCCGGCGACGAGACUGGUCAGAGCUGGGAUCUAAUCUCAA ..(((.((..((.((((.....(((((((..(((...(((((...)))))...)))...)))))))(((((((....))))))).....)))).))..))..)))(((((...))))).. ( -35.80) >DroAna_CAF1 6978 120 + 1 AGGGAGUUCUCAACCUCAUCCUGGAGGCUAUUGACAAGAUCAAUAUAAUUACAUCUCAGGGCUUCCUGGCCAGCUUCCUGGCCGGAGACGAGACUGGCCAAAGCUGGGACCUCAUCUCCA ((((((((((...((((......)))).((((((.....))))))............))))))))))((((((....))))))(((((.(((.(..((....))..)...))).))))). ( -47.60) >consensus AGGGCGUGCUCAAUCUCAUACUGGAGGCCAUUGACAAGAUCAACAUAAUCACCUCGCAGGGCUUCCUGGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUCUCCA .(((....)))...........(((((((..(((...(((.......)))...)))...)))))))...((((((..((((((((........))))))))))))))............. (-31.24 = -29.33 + -1.91)
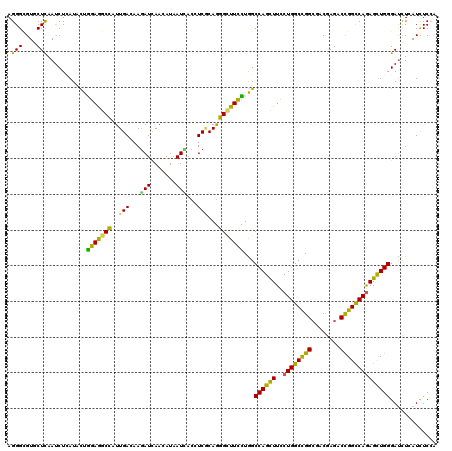
| Location | 4,380,281 – 4,380,401 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -30.78 |
| Energy contribution | -30.53 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
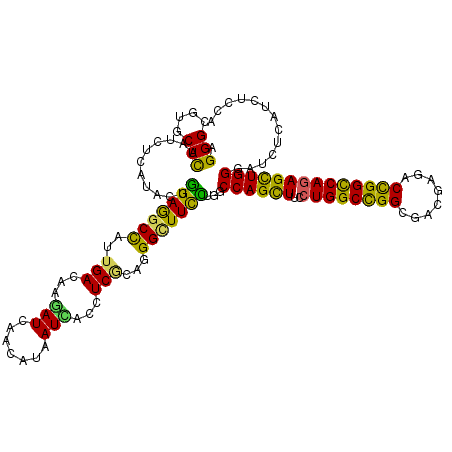
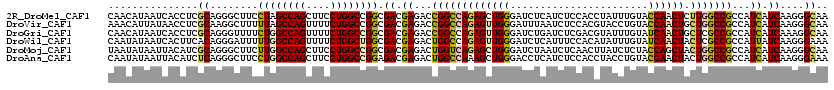
>2R_DroMel_CAF1 4380281 120 + 20766785 CAACAUAAUCACCUCGCAGGGCUUCCUAGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUCUCCACCUAUUUGUACCAACUCUUGGCCGCCAUCAUCAAGGGCAA ...........(((.(...(((.......((((((..(((((((((.....).)))))))))))))).......................(((.....)))..))).....).))).... ( -35.80) >DroVir_CAF1 60555 120 + 1 AAACAUUAUAACCUCGCAAGGCUUUUUAGCCAGUUUUCUGGCCGGCGACGAGACCGGCCAGAGUUGGGAUUUAAUCUCCACGUACCUGUACCAACUGCUGGCCGCCAUCAUCAAGGGCAA .............((....)).......((((((..((((((((((.....).)))))))))(((((((.......)))..(((....))))))).)))))).(((.........))).. ( -39.40) >DroGri_CAF1 7398 120 + 1 CAACAUAAUCACCUCGCAGGGUUUUCUGGCCAGUUUUCUGGCCGGCGACGAGACCGGCCAGAGUUGGGAUCUGAUCUCGACGUAUUUGUAUCAACUGCUCGCCGCCAUCAUCAAAGGCAA ..........((((....)))).....((((((((.((((((((((.....).)))))))))((((((((...))))))))...........)))))...)))(((.........))).. ( -40.40) >DroWil_CAF1 8161 120 + 1 CAAUAUAAUCACUUCACAGGGAUUUUUGGCCAGUUUUCUGGCUGGCGACGAGACUGGCCAGAGUUGGGAUCUCAUUUCCACAUAUUUGUAUCAACUACUCGCCGCCAUUAUCAAGGGAAA ............(((.(...(((....((((((....))))))((((.((((.((....))((((((((.......)))(((....)))..))))).)))).))))...)))...)))). ( -32.60) >DroMoj_CAF1 2719 120 + 1 UAAUAUAAUUACAUCGCAGGGCUUCUUGGCCAGCUUCCUGGCCGGCGACGAGACUGGUCAGAGCUGGGAUCUAAUCUCAACUUAUCUCUACCAGCUACUGGCCGCCAUCAUCAAGGGCAA ...........(.(((((((....)))((((((....)))))).)))).).....((((((((((((.......................)))))).))))))(((.........))).. ( -39.10) >DroAna_CAF1 7018 120 + 1 CAAUAUAAUUACAUCUCAGGGCUUCCUGGCCAGCUUCCUGGCCGGAGACGAGACUGGCCAAAGCUGGGACCUCAUCUCCACCUACCUGUACCAACUACUGGCCGCCAUCAUCAAGGGAAA .............(((((((.......((((((....))))))(((((.(((.(..((....))..)...))).))))).))).((.(((.....))).)).............)))).. ( -36.90) >consensus CAACAUAAUCACCUCGCAGGGCUUCCUGGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUCUCCACCUAUCUGUACCAACUACUGGCCGCCAUCAUCAAGGGCAA ...............((........((((((((....)))))))).((.((...(((((((((((((.......................)))))).)))))))...)).))....)).. (-30.78 = -30.53 + -0.25)
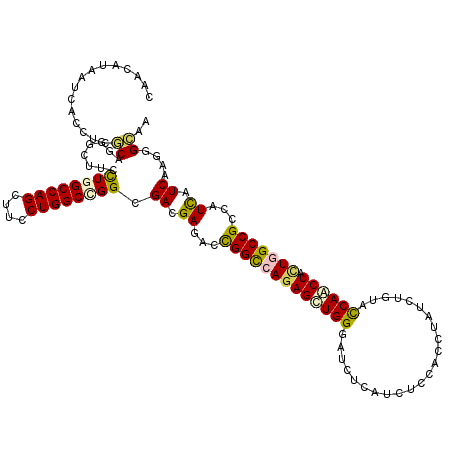
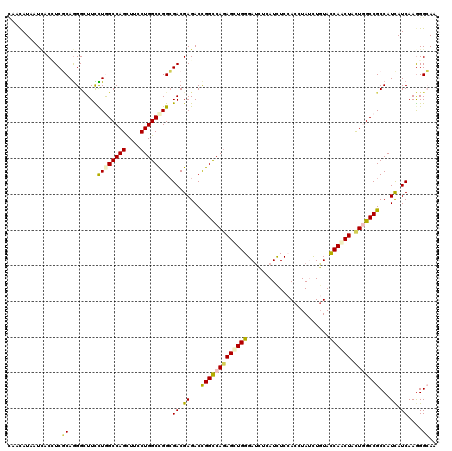
| Location | 4,380,281 – 4,380,401 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -32.90 |
| Energy contribution | -33.52 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
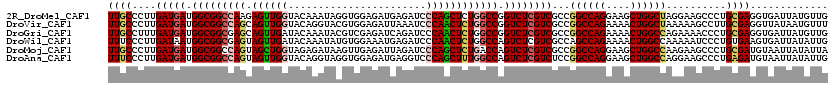
>2R_DroMel_CAF1 4380281 120 - 20766785 UUGCCCUUGAUGAUGGCGGCCAAGAGUUGGUACAAAUAGGUGGAGAUGAGAUCCCAGCUCUGGCCGGUCUCGUCGCCGGCCAGGAAGCUGGCUAGGAAGCCCUGCGAGGUGAUUAUGUUG (..(((..(((((.(.((((((.((((((((((......)))((.......))))))))))))))).).)))))((.((((((....))))))(((....)))))).))..)........ ( -46.60) >DroVir_CAF1 60555 120 - 1 UUGCCCUUGAUGAUGGCGGCCAGCAGUUGGUACAGGUACGUGGAGAUUAAAUCCCAACUCUGGCCGGUCUCGUCGCCGGCCAGAAAACUGGCUAAAAAGCCUUGCGAGGUUAUAAUGUUU ....(((((.....(((((((((..(((((((....)))..(((.......)))))))((((((((((......))))))))))...)))))).....)))...)))))........... ( -45.30) >DroGri_CAF1 7398 120 - 1 UUGCCUUUGAUGAUGGCGGCGAGCAGUUGAUACAAAUACGUCGAGAUCAGAUCCCAACUCUGGCCGGUCUCGUCGCCGGCCAGAAAACUGGCCAGAAAACCCUGCGAGGUGAUUAUGUUG (..((((.......((((((((((..(((((........)))))(..((((.......))))..).).)))))))))((((((....))))))............))))..)........ ( -42.30) >DroWil_CAF1 8161 120 - 1 UUUCCCUUGAUAAUGGCGGCGAGUAGUUGAUACAAAUAUGUGGAAAUGAGAUCCCAACUCUGGCCAGUCUCGUCGCCAGCCAGAAAACUGGCCAAAAAUCCCUGUGAAGUGAUUAUAUUG ..((.(((.(((.((((((((((..((....)).......(((....(((.......)))...)))..))))))))))(((((....)))))..........))).))).))........ ( -34.00) >DroMoj_CAF1 2719 120 - 1 UUGCCCUUGAUGAUGGCGGCCAGUAGCUGGUAGAGAUAAGUUGAGAUUAGAUCCCAGCUCUGACCAGUCUCGUCGCCGGCCAGGAAGCUGGCCAAGAAGCCCUGCGAUGUAAUUAUAUUA ((((....(((((.(((((.(((.((((((..((..(((.......)))..))))))))))).)).))))))))...((((((....))))))..........))))............. ( -41.50) >DroAna_CAF1 7018 120 - 1 UUUCCCUUGAUGAUGGCGGCCAGUAGUUGGUACAGGUAGGUGGAGAUGAGGUCCCAGCUUUGGCCAGUCUCGUCUCCGGCCAGGAAGCUGGCCAGGAAGCCCUGAGAUGUAAUUAUAUUG .........(((((.((((((((.((((((.((...((........))..)).)))))))))))).((((((..(((((((((....)))))).)))..)...))))))).))))).... ( -45.20) >consensus UUGCCCUUGAUGAUGGCGGCCAGUAGUUGGUACAAAUACGUGGAGAUGAGAUCCCAACUCUGGCCAGUCUCGUCGCCGGCCAGAAAACUGGCCAAAAAGCCCUGCGAGGUGAUUAUAUUG ((((....(((((.(((((((((.((((((.......................)))))))))))).))))))))...((((((....))))))..........))))............. (-32.90 = -33.52 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:10 2006