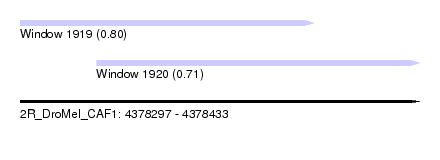
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,378,297 – 4,378,433 |
| Length | 136 |
| Max. P | 0.798406 |

| Location | 4,378,297 – 4,378,397 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -11.03 |
| Energy contribution | -12.12 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798406 |
| Prediction | RNA |
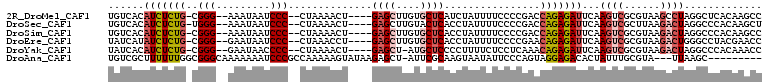
Download alignment: ClustalW | MAF
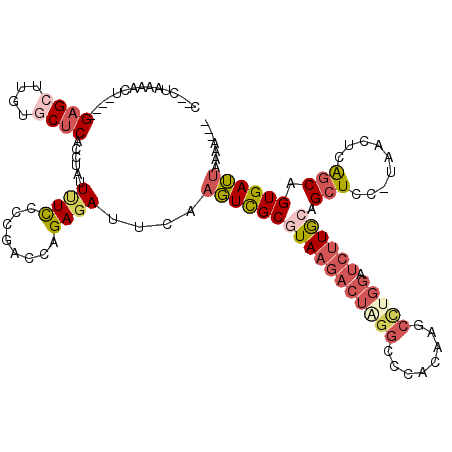
>2R_DroMel_CAF1 4378297 100 + 20766785 UGUCACAUCUCUG-CGGG--AAAUAAUCCC--CUAAAACU----GAGCUUGUGCUCAUCUAUUUUCCCCGACCAGAGAUUCAAGUCGCGUAAGCCUAGGCUCACAAGCC ......(((((((-.(((--(.....))))--.......(----((((....)))))...............))))))).......((....))...((((....)))) ( -26.30) >DroSec_CAF1 628 100 + 1 UGUCACAUCUCUG-UGGG--AAAUAAUCCC--CUAAAACU----GAGCUUGUACUCACCUAUUUUCCCCGACCAGAGAUUCAAGUCGCUUAAGACUAGGCCCACAAGCU .(((..(((((((-.(((--(.....))))--.......(----(((......))))...............)))))))...((((......)))).)))......... ( -21.40) >DroSim_CAF1 630 100 + 1 UGUCACAUCUCUG-CGGG--AAAUAAUCCC--CUAAAACU----GAGCUUGUGCUCACCUAUUUUCCCCGACCAGAGAUUCAAGUCGCGUAAGACUAGGCCCACAAGCC ......(((((((-.(((--(.....))))--.......(----((((....)))))...............)))))))...((((......)))).(((......))) ( -26.00) >DroEre_CAF1 629 100 + 1 UAUCAUAUCUCUG-CGGG--GAAUAAUCCC--CUAAACCU----GAGCUUGUGCUCACCUAUUUUCCCCGAACAGAGAUUCAAGUCGCGUAAGACUGGGCCUACGAACC ..((..(((((((-((((--(((.......--.......(----((((....)))))......)))))))..)))))))((.((((......)))).)).....))... ( -28.75) >DroYak_CAF1 627 99 + 1 UAUCACAUCUCUG-CGGG--GAAUAACCCC--CUAAAACU----GAGCU-AUGCUCCCCUUUUCUCCUCAAACAGAGAUUCAAGUCGCGUAAGACUAGGCCCACAAACC ......(((((((-.(((--(......)))--).......----((((.-..))))................)))))))((.((((......)))).)).......... ( -23.00) >DroAna_CAF1 5061 96 + 1 UGUCGCUUUUUUGGCGGGCAAAAAAAUCCCGCCAAAAAGUAUAAGAGCU-AUUCGCAAGUAAUAUUCCCAGUAGGAGACACUAUUUGCGUA---UUAAGC--------- ....(((((((.((((((.........)))))))))))))......(((-...((((((((...((((.....))))....))))))))..---...)))--------- ( -28.90) >consensus UGUCACAUCUCUG_CGGG__AAAUAAUCCC__CUAAAACU____GAGCUUGUGCUCACCUAUUUUCCCCGACCAGAGAUUCAAGUCGCGUAAGACUAGGCCCACAAGCC ......(((((((..(((.........)))..............((((....))))................)))))))...((((......))))............. (-11.03 = -12.12 + 1.08)
| Location | 4,378,323 – 4,378,433 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -14.61 |
| Energy contribution | -16.27 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
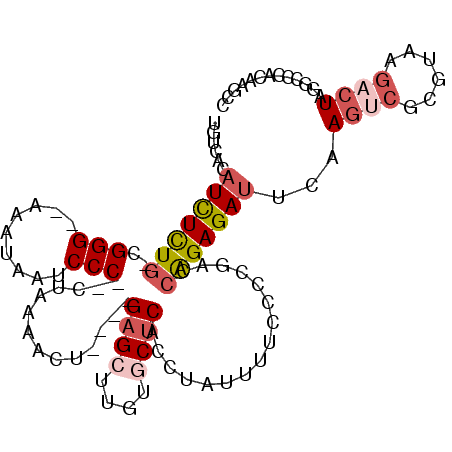
>2R_DroMel_CAF1 4378323 110 + 20766785 C--CUAAAACU----GAGCUUGUGCUCAUCUAUUUUCCCCGACCAGAGAUUCAAGUCGCGUAAGCCUAGGCUCACAAGCCUGGAUCUUGCAGCUCC-UAACUCAGCAGUGAUUAAAA--- .--.......(----((((....))))).........................(((((((((((.(((((((....)))))))..))))).(((..-......))).))))))....--- ( -29.50) >DroSec_CAF1 654 110 + 1 C--CUAAAACU----GAGCUUGUACUCACCUAUUUUCCCCGACCAGAGAUUCAAGUCGCUUAAGACUAGGCCCACAAGCUUGGAUCUUGCAGCUCC-UAACUCAGCAGUGAUUAAAA--- .--......(.----.(((((((.....((((((((...((((..(.....)..))))...)))).))))...)))))))..)...(..(.(((..-......))).)..)......--- ( -23.30) >DroSim_CAF1 656 110 + 1 C--CUAAAACU----GAGCUUGUGCUCACCUAUUUUCCCCGACCAGAGAUUCAAGUCGCGUAAGACUAGGCCCACAAGCCUGGAUCUUGCAGCUCC-UAACUCAGCAGUGAUUAAAA--- .--.......(----((((....))))).........................((((((((((((((((((......)))))).)))))).(((..-......))).))))))....--- ( -31.30) >DroEre_CAF1 655 110 + 1 C--CUAAACCU----GAGCUUGUGCUCACCUAUUUUCCCCGAACAGAGAUUCAAGUCGCGUAAGACUGGGCCUACGAACCUGGAUCUUGCAGCUCC-UAACUCAGCAGUGAUUAAAA--- .--.......(----((((....))))).........................(((((((((((((..((........))..).)))))).(((..-......))).))))))....--- ( -26.80) >DroYak_CAF1 653 109 + 1 C--CUAAAACU----GAGCU-AUGCUCCCCUUUUCUCCUCAAACAGAGAUUCAAGUCGCGUAAGACUAGGCCCACAAACCAGGAUCUUACAGCUCC-UAACUCAGCAGUGAUAUAAA--- .--........----((((.-..))))......((((........)))).....(((((((((((((.((........)).)).)))))).(((..-......))).))))).....--- ( -22.10) >DroAna_CAF1 5090 102 + 1 CGCCAAAAAGUAUAAGAGCU-AUUCGCAAGUAAUAUUCCCAGUAGGAGACACUAUUUGCGUA---UUAAGC--------------CUUAAAGCUCUUUAACUAGACAGUGAAUUAAACAA (((.....(((..(((((((-((.((((((((...((((.....))))....)))))))).)---).....--------------.....)))))))..))).....))).......... ( -20.00) >consensus C__CUAAAACU____GAGCUUGUGCUCACCUAUUUUCCCCGACCAGAGAUUCAAGUCGCGUAAGACUAGGCCCACAAGCCUGGAUCUUGCAGCUCC_UAACUCAGCAGUGAUUAAAA___ ...............((((....))))......((((........))))....(((((((((((((((((........))))).)))))).(((.........))).))))))....... (-14.61 = -16.27 + 1.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:07 2006