| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,375,782 – 4,375,919 |
| Length | 137 |
| Max. P | 0.993540 |

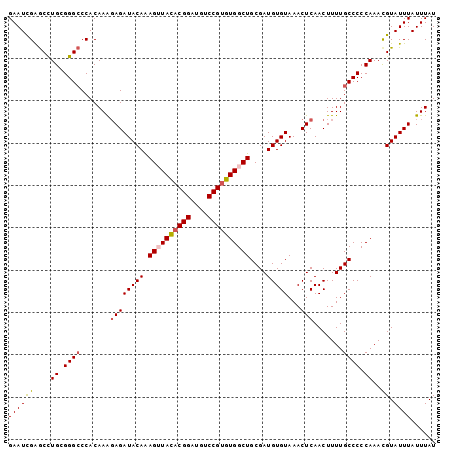
| Location | 4,375,782 – 4,375,883 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 93.96 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
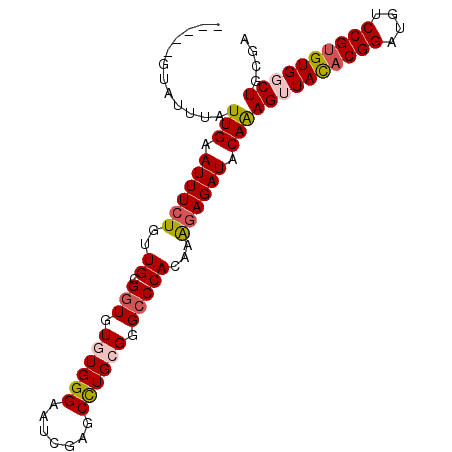
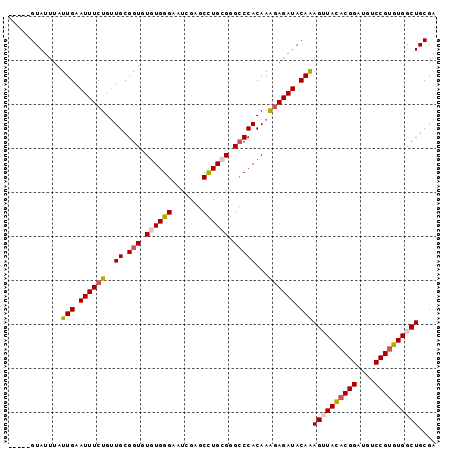
>2R_DroMel_CAF1 4375782 101 - 20766785 GAAUCGAGCCUGCGGGCCCACAAAGAGAUACAAAGUUACACGGAUGUCCGUGUGGCUGCGAUGUGUAAACUCAACUUUUGCCCCCAAACGUAUUUAUUUAU ((((((....((.((((.......((((((((.((((((((((....))))))))))....)))))...))).......)))).))..)).))))...... ( -28.24) >DroSec_CAF1 46430 101 - 1 GAAUUGAGCCUGCGGGCCCACAAAGAGAUACAAAGUUACACGGAUGUCCGUGUGGCUGCGAUGUGUAAACUCAACUUUUGCCCCCAAACGUAUUUAUUUAU ((((...(..((.((((.......((((((((.((((((((((....))))))))))....)))))...))).......)))).))..)..))))...... ( -28.04) >DroSim_CAF1 46636 101 - 1 GAAUUGAGCCUGCGGGCCCACAAAGAGAUACAAAGUUACACGGAUGUCCGUGUGGCUGCGAUGUGUAAACUCAACUUUUGCCCCCAAACGUAUUUAUUUAU ((((...(..((.((((.......((((((((.((((((((((....))))))))))....)))))...))).......)))).))..)..))))...... ( -28.04) >DroEre_CAF1 43079 100 - 1 GAAUCGGACUUGGGGG-CCACAAGCAGAUACAAAGAUACACGGAUGUCCGUGUGGCUGCGAUGUGUAAACUCAACUUUUGCCCCCAAACGUAUUUAUUUAU ((((((...(((((((-(((((.((((.........(((((((....))))))).))))..))))..............)))))))).)).))))...... ( -34.43) >DroYak_CAF1 44897 101 - 1 GAAUCGGGCCUGAGGGCCCACAAAGAGAUACAGAGAUAUUCGGAUGUCCGUGUGGCUGCGAUGUGUAAACUCAACUUUUGCCCCCAAAUGUAUUUAUUUAU .....(((((....)))))......(((((((........(((....)))..(((..((((..((......))....))))..)))..)))))))...... ( -26.00) >consensus GAAUCGAGCCUGCGGGCCCACAAAGAGAUACAAAGUUACACGGAUGUCCGUGUGGCUGCGAUGUGUAAACUCAACUUUUGCCCCCAAACGUAUUUAUUUAU ((((((....((.((((.......((((((((.((((((((((....))))))))))....)))))...))).......)))).))..)).))))...... (-23.38 = -23.82 + 0.44)


| Location | 4,375,822 – 4,375,919 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -17.37 |
| Energy contribution | -17.69 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4375822 97 + 20766785 UCGCAGCCACACGGACAUCCGUGUAACUUUGUAUCUCUUUGUGGGCCCGCAGGCUCGAUUCCCACACACCGCAACAGAAAUUCAAUAAAUACCGAAU (((.((((((((((....))))))...............((((....)))))))))))....................................... ( -22.60) >DroSec_CAF1 46470 97 + 1 UCGCAGCCACACGGACAUCCGUGUAACUUUGUAUCUCUUUGUGGGCCCGCAGGCUCAAUUCCCACACACCGCAUCAGAAAUUCAAUAAAUACCGAAU ..((....((((((....)))))).....(((.........((((((....))))))........)))..))......................... ( -20.43) >DroSim_CAF1 46676 92 + 1 UCGCAGCCACACGGACAUCCGUGUAACUUUGUAUCUCUUUGUGGGCCCGCAGGCUCAAUUCCCACACACCGCAACAGAAAUUCAAUAAAUAC----- ..((....((((((....)))))).....(((.........((((((....))))))........)))..))....................----- ( -20.43) >DroEre_CAF1 43119 91 + 1 UCGCAGCCACACGGACAUCCGUGUAUCUUUGUAUCUGCUUGUGG-CCCCCAAGUCCGAUUCCCACACACCGCAACAGAAAUUCAAUAAAUAC----- ..((((..((((((....))))))....)))).((((.((((((-.......................))))))))))..............----- ( -19.50) >DroYak_CAF1 44937 92 + 1 UCGCAGCCACACGGACAUCCGAAUAUCUCUGUAUCUCUUUGUGGGCCCUCAGGCCCGAUUCCCACACACCGCAACAGAAAUUCAAUAAAUAC----- ..((((.....(((....))).......))))...((((((((((((....)))))(.....).......)))).)))..............----- ( -17.40) >consensus UCGCAGCCACACGGACAUCCGUGUAACUUUGUAUCUCUUUGUGGGCCCGCAGGCUCGAUUCCCACACACCGCAACAGAAAUUCAAUAAAUAC_____ ..((....((((((....)))))).....(((.........((((((....))))))........)))..))......................... (-17.37 = -17.69 + 0.32)

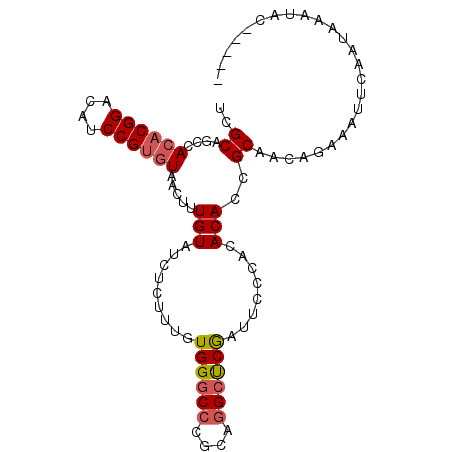
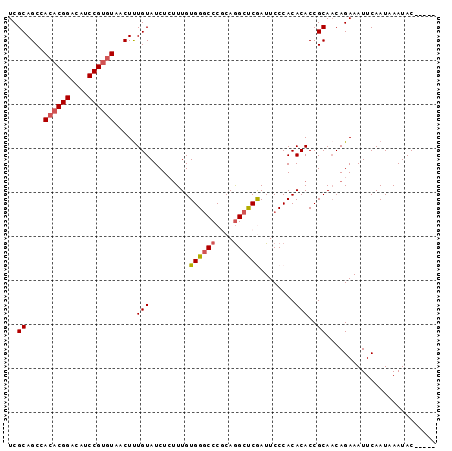
| Location | 4,375,822 – 4,375,919 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -27.66 |
| Energy contribution | -28.42 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4375822 97 - 20766785 AUUCGGUAUUUAUUGAAUUUCUGUUGCGGUGUGUGGGAAUCGAGCCUGCGGGCCCACAAAGAGAUACAAAGUUACACGGAUGUCCGUGUGGCUGCGA ..(((.......(((.((((((..((.(((.((..((.......))..)).)))))...)))))).)))((((((((((....)))))))))).))) ( -36.40) >DroSec_CAF1 46470 97 - 1 AUUCGGUAUUUAUUGAAUUUCUGAUGCGGUGUGUGGGAAUUGAGCCUGCGGGCCCACAAAGAGAUACAAAGUUACACGGAUGUCCGUGUGGCUGCGA ..(((.......(((.((((((..((.(((.((..((.......))..)).)))))...)))))).)))((((((((((....)))))))))).))) ( -36.40) >DroSim_CAF1 46676 92 - 1 -----GUAUUUAUUGAAUUUCUGUUGCGGUGUGUGGGAAUUGAGCCUGCGGGCCCACAAAGAGAUACAAAGUUACACGGAUGUCCGUGUGGCUGCGA -----.......(((.((((((..((.(((.((..((.......))..)).)))))...)))))).)))((((((((((....)))))))))).... ( -35.00) >DroEre_CAF1 43119 91 - 1 -----GUAUUUAUUGAAUUUCUGUUGCGGUGUGUGGGAAUCGGACUUGGGGG-CCACAAGCAGAUACAAAGAUACACGGAUGUCCGUGUGGCUGCGA -----.......(((....(((((((.(((.(.((((.......)))).).)-)).).))))))..)))((.(((((((....))))))).)).... ( -24.30) >DroYak_CAF1 44937 92 - 1 -----GUAUUUAUUGAAUUUCUGUUGCGGUGUGUGGGAAUCGGGCCUGAGGGCCCACAAAGAGAUACAGAGAUAUUCGGAUGUCCGUGUGGCUGCGA -----(((.((((...((((((((.......((((((..(((....)))...)))))).......))))))))...(((....))).)))).))).. ( -25.84) >consensus _____GUAUUUAUUGAAUUUCUGUUGCGGUGUGUGGGAAUCGAGCCUGCGGGCCCACAAAGAGAUACAAAGUUACACGGAUGUCCGUGUGGCUGCGA ............(((.((((((..((.(((.((((((.......)))))).)))))...)))))).)))((((((((((....)))))))))).... (-27.66 = -28.42 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:04 2006